Word file (46 KB )
advertisement

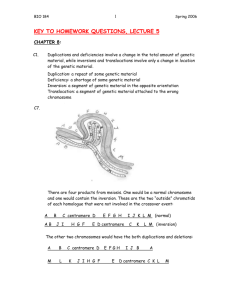
SUPPLEMENTARY MATERIAL Strategy for engineering the translocations into S.cerevisiae In order to generate the reciprocal chromosomal translocation, the FY23 (Kanr, Ura+) strain was transformed with the plasmid Yep351-cre-cyh, which contains the Cre recombinase gene, with the Candida maltosa cycloheximide-resistance gene, and the S. cerevisiae LEU2 gene as selectable markers1. Transformants were selected on SD Leu- minimal medium plates, and growing the cells in YPGal medium induced the Cre recombinase. The selectable markers, at the two sites, were excised and, in a proportion of the survivors, centromere-distal fragments were exchanged between chromosomes VI and VII to generate a reciprocal chromosomal translocation (VItVII and VIItVI). Survivors were screened for those bearing the required translocation by diagnostic PCR, using oligonucleotides annealing to regions flanking either side of the putative translocation breakpoint on chromosome VI or VII (see Fig.1 in paper). The change in the karyotype was verified by CHEF analysis and the strain, bearing the required translocation, was designated Sct1. Sct1 was then manipulated to generate a second translocation between the right arm of chromosome VI (which comprises the genetic material of chromosome VII, because of the translocation introduced in Sct1) and the left arm of chromosome XVI, in order to render it colinear with the second S. mikatae strain, IFO1815. For this construct, the translocation breakpoints were located between ORFs YGR188c and YGR189c, and YPL103c and YPL116w2. In the case of the last breakpoint, the translocation falls in a rather large chromosomal interval (ca 30 kb) and no genomic sequence is available for 1815 so (for technical reasons) we chose the largest intergenic region, between ORFs YPL108w and YPL109c, to insert the loxP cassette. This generated a new strain, Sct1/2, which is colinear to S. mikatae IFO1815. During the screen for Sct1/2, colonies carrying the second translocation (t2) but which had lost the original translocation (t1) from strain Sct1 - due to a back-translocation between chromosomes VI and VII - were also isolated. 1. Delneri, D. et al. Exploring redundancy in the yeast genome: an improved strategy for the use of the cre-loxP system. Gene 252, 127-135 (2000). 2. Fischer, G. et al. Chromosomal evolution in Saccharomyces. Nature 405, 451-454 (2000). Table A: Oligonucleotides used for the construction of the cassettes used for creating the translocation between chromosome VI and VII, and between chromosome VII and XVI. Name Sequencea Template YFR06-07F 5’-atgattactagtgcttaaaccttgttattcatgcctcagctgaagcttcgtacgc-3’ PUG6 YFR06-07R 5’-aacctaacaaggttacttcatttttcccgaagagcaatacgcataggccactagtggatc-3’ PUG6 YGR21-22F 5’-actgtaataaagagtaaaaacaacaacaaagacaatcgagctgaagcttcgtacgc-3’ PUGKlURA3 YGR21-22R 5’-gtgtttaaacaaatttcggaagcaatggagcataggccactagtggatc-3’ PUGKlURA3 Ck6F 5’-cgtcgtccacctatttgag-3’ Genomic DNA Ck6R 5’-atccaatagctgaagaacgtc-3’ Genomic DNA KanMX-R 5’-caatcgatagattgtcgcac-3’ Genomic DNA Ck7F 5’-acatcagagacttgggcatc-3’ Genomic DNA Ck7R 5’-agctgtgctcgatgttgtg-3’ Genomic DNA URA-R 5’-ctcatcagtcgaacgaacgt-3’ Genomic DNA YGR188-189F 5’- ccgcactttatattttaacgtttgatggttttctatcttggcagcgcagctgaagcttcgtacg-3’ PUG6 YGR188-189R 5’- cttttattattaagggtagccttcatacaaaggaaatttatgtcatcgcataggccactagtggatc-3’ PUG6 YPL116-103F 5’-cgctgcttctataacacttgttttgacatgacgataagtgtagtcagctgaagcttcgtacgc-3’ PUG-KlURA3 YPL116-103R 5’-ataaattgattctaagtacaggaagtatcaagggggggggttgacgcataggccactagtggatc-3’ PUG-KlURA3 Ck7bF 5’-gttggcacaagttatggagtc-3’ Genomic DNA Ck7bR 5’-cgtagctttactagccttgc-3’ Genomic DNA Ck16F 5’-ttattcccctcatatattcgg-3’ Genomic DNA Ck16R 5’-ttattatgcaagttgtgtaggc-3’ Genomic DNA a the underlined bases are the sequences complementary to the templates. Tanslocation t1: the breakpoint on chromosome VI was located between ORF YFR006 and YFR007 (where a tRNA TF(GAA)F and an LTR were present; another tRNA TG (GCC) F1 and LTR were also present between YFR008w and YFR009w); the breakpoint on chromosome VII was located, for technical reasons, between ORF YGR021w and YGR022w (the only tRNA TD(GUC)G1 was present in between ORF YGR022c/YGR023w and YGR024c). Following the recent release of the sequence of the S. mikatae 1816 (http://genomewww4.stanford.edu/cgi-bin/FUNGI/FungiMap), it is now confirmed that the exact breakpoints are between ORF YFR008w and YFR009w for chromosome VI and between ORF YGR022c/YGR023w and YGR024c for chromosome VII. Translocation t2: the breakpoint on chromosome VII was mapped precisely between ORF YGR188c and YGR189c (where a tRNA TK(CUU)G3 and an LTR were present); the breakpoint on chromosome XVI was placed, again for technical reasons, between ORF YPL108wand YPL109c (only one tRNA TM(CAU)P was present in the breakpoint interval between ORF YPL111w and YPL112c). No sequence is available for S. mikatae 1815. Table B: Oligonucleotides used for the generation of species-specific chromosomal probes. Chromosomes I II III IV V VI VII VIII IX X XI XII XIII XIV XV XVI Primers specific for S. cerevisiae Primers specific for S. mikatae 1CF: 5'-GTTGGCTAGTTTCGCATTC-3' 1MF: 5'-TGACTAGCCGTTGGATGTAC-3' 1CR: 5'-CCTTCATTGTTCTTGTCTGATG-3' 1MR: 5'-CAACGACAGATTTCGAGTACC-3' 2CF: 5'-TGCGGTAGCATTTCCATC-3' 2MF: 5'-CTAATTAAATCATTAGTGGGGC-3' 2CR: 5'-GGTTAGTGGCCAGATTGAGA-3' 2MF: 5'-GATGGCAATAGATTACCTATGC-3' 3CF: 5'-ACCTTTCCCTGATGTACGTC-3' 3MF: 5'-TGATATACGGTACAAGAAAGGG-3' 3CR: 5'-CTTCACATCAAACTTGTGCC-3' 3MR: 5'-AAGCTCAAACAAATCCAATG-3' 4CF: 5'-GACAAGCAACACAGTGTTTTAG-3' 4MF: 5'-GCAAATAATTCTTATGGCCC-3' 4CR: 5'-AGCTACTGTGACGACCAGG-3' 4MR: 5'-TGGAGGAACAACATTACAAAAC-3' 5CF: 5'-ACGACATTCCATCTCATCG-3' 5MF: 5'-CTTCTCATAAATAGGCTTGGC-3' 5CR: 5'-GAGAGCGTGAGAAAATACTGC-3' 5MR: 5'-GTTTTACAAAGAAAGCGTGC-3' 6CF: 5'-TTAGCAACTCTATGTACAACCG-3' 6MF: 5'-TGAAATAACTGACTGTCGTTTG-3' 6CR: 5'-TGATAAGTTTGATTGCGTCC-3' 6MR: 5'-AGGAACATGCCATTCAAAG-3' 7CF: 5'-GAATCTCTTGGTAGACTCGACC-3' 7MF: 5'-GCAAAAATCTATAATACTGCTCG-3' 7CR: 5'-GAAATGAACCTGCCAGAAG-3' 7MR: 5'-CTACGGATACTGGTGAAATCG-3' 8CF: 5'-ATAGAATAGTCCCATGGCG-3' 8MF: 5'-TTGTCCTTTTGTCAGTATGTTG-3' 8CR: 5'-CATGGGAGTCAGTTCTTGTTC-3' 8MR: 5'-TTTATCATTTATCTCGTAGGACC-3' 9CF: 5'-ATAATGGCAATTGTGGAATG-3' 9MF: 5'-CTAGAACATCCTCCGGAATC-3' 9CR: 5'-TTACCAGGCGTACTATTTGG-3' 9MR: 5'-GGGTAGACTCCCTAAGTGTTG-3' 10CF: 5'-CGAATCAAGAACCTTGGTG-3' 10MF: 5'-TCCATGGAGCTTAATAGCG-3' 10CR: 5'-AAACGTCGCAGATATGGAC-3' 10MR: 5'-AACTTTGAAGCATCCTTTGAC-3' 11CF: 5'-GCACCATCCTTTAACTCCAC-3' 11MF: 5'-AGATTGAAGTTCGGATACGTG-3' 11CR: 5'-GAAAAAGATGCACTCTGTCG-3' 11MR: 5'-TGATCAGCTAGCACATATTGC-3' 12CF: 5'-TGAGTCGTCATCACCATACG-3' 12MF: 5'-TTTCACAGTCGTCTCGATTG-3' 12CR: 5'-AGATTTTGTCCAAAGTTAGCAG-3' 12MR: 5'-CAATTCGGGTTTCCATAAC-3' 13CF: 5'-GTCATCCTGACATTGCTTTC-3' 13MF: 5'-CACAGTCATAGGTGAACTGAGG-3' 13CR: 5'-ACGATGAGAGAGGAGAAACG-3' 13MR: 5'-TTCGGTAAAAACATCCTGG-3' 14CF: 5'-GGTGAAGAAAAGTTTGTAAAATG-3' 14MF: 5'-AAGAGTGCAATGTTACGGG-3' 14CR: 5'-TCGACCTCGATCTACTTCG-3' 14MR: 5'-TAAGTCATGGCAGGTCGG-3' 15CF: 5'-TGTTAAACTACAATTTCGGATTC-3' 15MF: 5'-GCTCATTTCTTTACTTGCTTG-3' 15CR: 5'-CACTGCCATTTTCATGACC-3' 15MR: 5'-ACGTAGAGCTCAAGTAGACGAC-3' 16CF: 5'-GTCACACCAGGCTATGAGG-3' 16MF: 5'-GTCGCCATATAAATGAGATAGC-3' 16CR: 5'-TCCACCTGTTACCTTCTTGAT-3' 16MR: 5'-ACATACAGAGAAGAATACCTTGC-3'