Supplementary data
advertisement

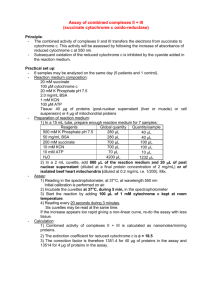
Supplementary Information Materials and Methods Cell culture L929sAhFas cells are L929sA cells transfected with cDNA of the human Fas receptor as described previously 1. The cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Paisly, Scotland) supplemented with 10% fetal calf serum (Greiner, Poitiers, France), 100 IU/ml penicillin (Gibco BRL, Paisley, Scotland), 0.1 mg/ml streptomycin (Gibco BRL) and 0.03% L-glutamine (Gibco BRL). Cytokines and reagents Recombinant murine TNF (6x107 IU/ml) and recombinant murine IFN-β (3x106 IU/ml) were produced in E. coli and purified in our laboratory. LiCl (ICN Biomedicals Inc, Ohio, USA) was dissolved at 1 M in medium. Polyinosinic-polycytidylic acid (poly(I)poly(C)), a synthetic dsRNA (Amersham Pharmacia Biotech, Rainham, UK), was dissolved at 3.5 mg/ml in water. Propidium iodide (PI; Becton Dickinson, Sunnyvale, CA, USA) was dissolved at 3 mM in PBS. Dihydrorhodamine 123 (DHR123; Molecular Probes, Eugene, OR, USA) was dissolved at 1 mM in DMSO. NADH (Boehringer Mannheim) was dissolved at 1 mM in water. The following products were purchased from Sigma Chemical Co. (St. Louis, MO) and dissolved as indicated: BHA and BHT (100 mM in ethanol), acetylCoA (3 mM in water), bovine serum albumin (BSA, 1% in water), malonate (0.5 M in water), KCN (30 mM in water), rotenone (5 mM in ethanol), antimycine A (0.1 mM in DMSO/water 1:2 v/v), horse cytochrome c (50 mg/ml in 10 mM potassium phosphate buffer, PPB, pH 7.0), 2,6dichloroindophenol sodium salt hydrate (DCPIP, 1 mM in 10 mM PPB, pH 7.5), sodium succinate (200 mM in 100 mM PPB, pH 7.5), 5,5’-dithiobis (2-nitrobenzoic acid) (DTNB, 1 mM in 0.1 M Tris-HCl, pH 8.1), oxaloacetic acid (10 mM in 0.1 M Tris-HCl, pH 8.1), decylubiquinone (65 mM in ethanol), AA861 (50 mM in ethanol) and BEL (80 mM in DMSO). MAFP (Biomol) was dissolved at 50 mM in ethanol and NDGA (Calbiochem) at 100 mM in ethanol. NaBH4 was purchased from Aldrich and heparin from B. Braun Melsungen AG (50 U/ml stock). Induction of necrosis for fluorescence-activated cell sorter (FACS) analysis L929sAhFas cells were seeded at 2x105 cells per well (uncoated 24-well tissue culture plates, Sarstedt Inc., Newton, NC) and incubated overnight in suspension. Following 24 h culture at 37°C, cells either received no treatment or were treated with murine IFN-β (200 IU/ml) 2. After another 24 h, the cells were pre-incubated for 0.5 h with different inhibitors or medium as indicated in the figure legends, and then stimulated with murine TNF (10 000 IU/ml) in combination with 10 mM LiCl 3, or dsRNA (2.5 µg/ml). MAFP was used at 100 μM, AA861 at 50 μM, NDGA at 100 μM, BEL at 30 μM, rotenone at 25 μM, malonate at 5 mM and antimycin A at 50 μM. Measurement of cell membrane permeability, hypoploidy and ROS production by FACS analysis Loss of cell membrane integrity as a measure of cell death was determined by PI fluorescence. The percentage of cells containing hypoploid DNA was determined by PI staining following one freeze-thaw cycle to permeabilize cells. PI was used at 30 µM and fluorescence was detected at 610 nm on a FACScalibur flow cytometer (Becton Dickinson Biosciences) equipped with a 488 nm argon ion laser. Production of ROS was measured at 525 nm and calculated as mean rhodamine 123 (R123) fluorescence resulting from oxidation of DHR123 by PI-negative cells. Cells were incubated with 0.1 µM DHR123 for 30 min before analysis. Relative rhodamine 123 fluorescence is defined as the ratio between fluorescence at a given time point and the initial fluorescence for the same condition. Preparation of muscle homogenates Skeletal muscle was isolated from C57BL/6 mice. Twenty volumes of homogenization buffer (10 mM Tris-HCl, 0.25 M sucrose, 2 mM EDTA, 50 U/ml heparin, pH 7.4) were added to one volume of tissue sample (50 mg) for homogenization in a glass/glass pestle. The homogenate was centrifuged (5 600 g) for 1 min. The supernatant (whole tissue homogenate) was kept on ice and used for spectrophotometric analysis. Protein concentrations were in the range of 3-5 mg/ml. Immediately before complex activity measurements, the homogenate was sonicated to disrupt mitochondrial membrane integrity and allow uptake of essential substrates. Spectrophotometric analysis of electron transport chain complex activities Spectrophotometric assays were performed to measure the activities of NADH-CoQ reductase (complex I), succinate CoQ oxidoreductase (complex II), cytochrome c-ubiquinole reductase (complex III), cytochrome c oxidase (complex IV) and citrate synthase, as previously described 4. For each experiment, the results are presented as the mean activity of the complex in 10 independent skeletal muscle samples, with error bars to indicate standard error of the mean (SEM). Briefly, each assay was set up as follows: NADH-CoQ reductase activity (complex I): A 50 µl sample of skeletal muscle homogenate was incubated in 25 mM potassium phosphate buffer (PPB), pH 7.4, containing 2.5 mg/ml BSA, 5 mM MgCl2, 100 µM NADH (substrate complex I), and 0.6 mM KCN (to block oxidation by complex IV). Inhibitors were added where appropriate. Treatment with 10 µM rotenone was used as a positive control of inhibition. After stirring and stabilization for 1 min at 30 °C, decylubiquinone (commercial quinone substitute) was added to a final concentration of 130 µM, and the rate of NADH oxidation at 340 nm (ε = 6 290 M-1cm-1) was integrated over consecutive intervals of 16 seconds for a period of 4 min. Results are corrected for mitochondrial input as determined by citrate synthase activity. Succinate CoQ oxidoreductase activity (complex II): A 20 µl sample of skeletal muscle homogenate was incubated in 10 mM PPB, pH 7.4, containing 20 mM sodium succinate (substrate for complex II) and 3 mM KCN (to block oxidation by complex IV), with or without inhibitors. Treatment with 5 mM malonate was used as a positive control of inhibition. After stirring and stabilization for 5 min at 37 °C, DCPIP (50 µM final) and decylubiquinone (130 µM final) were added. DCPIP reduction was determined at 600 nm (ε = 19 100 M-1cm-1) over consecutive intervals of 16 seconds for a period of 4 min. Results are corrected for mitochondrial input as determined by citrate synthase activity. Cytochrome c-ubiquinole reductase activity (complex III): A mixture of 900 µl of complex III buffer (31 mM KH2PO4, 3 mM EDTA, 0.6 mM KCN in DMSO/water (1:9 v/v), pH 7.5), 10 µl decylubiquinole (synthesized by reducing 13 mM decylubiquinone with NaBH4 5) and 100 µl cytochrome c (1% in 10 mM PPB, pH 7.0), with or without inhibitors, was stabilized for 3 min at 37 °C. Afterwards, a 10 µl sample of skeletal muscle homogenate was added. Treatment with 1 µM antimycin A was used as a positive control of inhibition. Reduction of cytochrome c was monitored at 550 nm (ε = 18 500 M-1cm-1) over consecutive intervals of 16 seconds for a period of 4 min. Results are corrected for mitochondrial input as determined by citrate synthase activity. Cytochrome c oxidase activity (complex IV): A 990 µl mixture of 10 mM PPB, pH 7.0, containing 0.1% reduced cytochrome c (stock solution of 1% in 10 mM PPB pH 7.0 reduced with sodium dithionit) was incubated for 10 min at 37 °C, with or without inhibitors. Afterwards, a 10 µl sample of skeletal muscle homogenate was added. Treatment with 0.6 mM KCN was used as a positive control for inhibition of complex IV. Oxidation of cytochrome c was monitored at 550 nm (ε = 18 500 M-1cm-1) over consecutive intervals of 30 seconds for a period of 5 min. Results are corrected for mitochondrial input as determined by citrate synthase activity. Citrate synthase activity: A 10 µl sample of skeletal muscle homogenate was incubated in 0.1 mM Tris-HCl buffer, pH 8.1, containing 60 µM acetylCoA, 100 µM DTNB and 0.05% Triton X-100. After stirring and stabilization for 2 min at 37 °C, oxaloacetic acid was added (0.5 mM final). Reaction with DTNB was measured at 412 nm (ε = 13 600 M-1cm-1) over consecutive intervals of 33 seconds for a period of 4 min 6. 1. Vercammen D et al. (1998) J Exp Med 188:(5): 919-30 2. Kalai M et al. (2002) Cell Death Differ 9:(9): 981-94 3. Beyaert R et al. (1989) Proc Natl Acad Sci U S A 86:(23): 9494-8 4. Van Coster R et al. (2001) Pediatric Research 50:(5): 1-8 5. Krahenbuhl S et al. (1994) Clin Chim Acta 230:(2): 177-87 6. Ellman G and Lysko H (1979) Anal Biochem 93:(1): 98-102 Supplementary figure 1: Structure of BHA and BHT