Taxonomy and Classification - Lin
advertisement

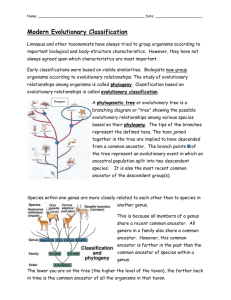
Taxonomy and Classification Taxonomy is the science dealing with description, identification, nomenclature, and classification of living things. A dichotomous key is a tool that allows users to identify items or organisms in a systematic and reproducible fashion. Dichotomous keys may be used in a variety of situations, such as for identifying rocks and minerals as well as for identifying unknown organisms to some taxonomic level (e.g., species, genus, family, etc.). What makes these keys distinctive is that they are ordered in such a way that a series of choices is made that leads the user to the correct identity of the item they are looking at. "Dichotomous" means, "divided into two parts." Therefore, dichotomous keys always offer two choices for each step, each of which describes key characteristics of a particular organism or group of organisms. Imagine that you are trying to construct a dichotomous key for the students in your biology laboratory class. To do this you would need to make a list of some characteristics possessed by the members of the group, such as gender, hair color, height, type of clothing (jeans, dress pants, shorts, dress), whether or not they wear glasses, and so forth. You would then construct your key by setting up a series of bifurcating characteristics, starting with the most general characters and moving to the most specific such that, in the end, each member of the group can be clearly identified. For example: 1. Sex female---2 Sex male---5 2. Hair color red---Sally Hair color not red---3 3. Hair color blonde---Julie Hair color black---4 4. Glasses worn---Deanna &nbseanna Glasses not worn---Leslie 5. Shoes high-top sneakers---Joseph Shoes not high-tops---6 6. Hair color blonde--Michael Hair color brown--David Thus if we are looking at a male student with brown hair wearing cowboy boots we would start at step 1. Because he is male, we would be instructed to go to point 5. At point 5 we would establish that his shoes are not high-tops, and so be directed to step 6, where the fact that our specimen has brown hair would allow us to determine that we are looking at David. Exercise 1 Imagine that you meet a blonde male member of this class wearing teva sandals. Who is it? Imagine that his girl friend has black hair but doesn't wear glasses. Who is he dating? Exercise 2 Use the dichotomous key to the principle orders insects provided to determine the identity of the following organisms: Click here for specimen 1. Click here for specimen 2. Click here for specimen 3. Systematic Approaches to Phylogeny What is Systematics? Systematics is a discipline within biology whose goal is the determination of the evolutionary history and relationships among organisms (termed phylogeny) and then the use of that phylogeny in classifying organisms. To achieve that goal, a systematist will utilize evidence from a wide variety of sources including paleontology, embryology, morphology, behavior, and molecular biology. Over the last few centuries systematists have developed a number of different approaches for trying to show the relationships of organisms. As a result, different schemes have emerged for ordering these relationships into a classification scheme. The main approaches in the last 50 or so years may be classified as follows: Phenetic (or numerical taxonomy) Cladistic (or phylogenetic) Evolutionary (or synthetic) Phenetics In the "old days" (before the twentieth century), classification was largely carried out by "gentlemen" scientists who based their identifications of organisms upon a few "important" characteristics and a "gut feeling" developed during many years experience. Phenetic classification methods were invented by systematists who thought that this type of classification scheme was too subjective. These scientists argue that taxonomy should not be based on a few characteristics arbitrarily judged to be "important," but rather upon the degree of overall similarity between organisms. Thus they collect data on as many characteristics of the organisms being classified as possible. Each organism is then compared with every other for all characters measured, and the number of similarities (or differences) is calculated. The organisms are then clustered in such a way that the most similar are grouped close together and the more different ones are linked more distantly. Because of the enormous amount of data involved in this process, these calculations are usually made using specially programmed computers1. The taxonomic clusters (phenograms) that result from such an analysis do not necessarily reflect genetic similarity or evolutionary relatedness. Pheneticists would argue that this is OK, because nobody really knows the true evolutionary histories of these organisms and at least this system is objective. However, the lack of evolutionary significance in phenetics has meant that this system has had little impact on animal classification, and as a consequence interest in and use of phenetics has been declining in recent years. 1 The dependence of Phenetics on the use of computers made it particularly unpopular with some "old-school" biologists. Paul Ehrlich, one of the early converts to Phenetics, was reportedly once asked by an irate taxonomist at a conference, "You mean to tell me that taxonomists can be replaced by computers?" Ehrlich responded, "No, some of you can be replaced by an abacus!" Cladistics (Phylogenetic Systematics) Phenetics has been criticized because phenograms resulting from such analyses do not necessarily correspond to evolutionary histories (degree of relatedness) between organisms. Thus an alternative approach to diagramming relationships between taxa was developed, called cladistics. The basic assumption behind cladistics is that members of a group share a common evolutionary history, and are thus more "closely related" to one another than they are to other groups of organisms. Related groups of organisms are recognized because they share a set of unique features (apomorphies) which were not present in distant ancestors, but which are shared by most or all of the organisms within the group. These shared derived characteristics are called synapomorphies. In contrast to phenetics, in cladistics groupings do not depend on whether organisms share physical traits, but on their evolutionary relationships. Indeed, in cladistic analyses two organisms may share numerous characteristics but still be considered members of different groups. For example, a jellyfish, a sea star, and a human: jellyfish and sea stars both live in the ocean, have radial symmetry and are invertebrates, so phenetic analysis might place them together in a group. However, this would not reflect evolutionary relationships, as sea stars are actually more closely related to humans than they are to jellyfish (both are deuterostomes). Thus in cladistics, the emphasis is not upon the presence of all shared traits, but upon the presence of shared derived (apomorphic) traits. In the example above radial symmetry, aquatic habitat and invertebrate structure are all traits that are believed to have been present in the common ancestor of all animals, and so such traits are not considered to be very useful in determining relationships using cladistic analysis. Cladistic analyses have some pretty strict rules. For example, cladists always assume that new species arise by bifurcations of the original lineage (the lineage always splits in two). Most cladists assume that the original ancestral species no longer exists after this bifurcation, so each branching event results in two new species. In addition, cladistic groupings must possess the following characteristics: 1. All species in a grouping must share a common ancestor. 2. All species derived from a common ancestor must be included in the taxon. The application of these requirements results in the following terms being used to describe the different ways in which groupings can be made: A monophyletic grouping is one in which all species share a common ancestor and all species derived from that common ancestor are included. This is the only form of grouping accepted as valid by cladists. (For example, turtles, lizards, crocodilians and birds are all derived from a shared common ancestor. Thus a monophyletic grouping would place all of these together, rather than placing birds into a separate group.) A paraphyletic grouping is one in which all species share a common ancestor, but not all species derived from that common ancestor are included (for example, grouping turtles, lizards and crocodiles as "reptiles" and separating that grouping from the birds). A polyphyletic grouping is one in which species that do not share an immediate common ancestor are lumped together, while excluding other members that would link them (for example, a hypothetical group the "lizmams" made by grouping together the lizards and the mammals). Thus, in cladistics, no matter how divergent in appearance B might be from C, relative to A, if B and C share a common ancestor that is not shared by A, then B and C must be grouped together and separated from lineage A. In the cladogram of the reptiles and birds above, you can see an example of such a situation. Steps for Constructing a Cladogram 1. Select a taxonomic group to be analyzed; for example, a group of vertebrates. 2. For each member of the group, determine some observable traits (characters), and note their "states" (a "character state" is one of two [or more] possible forms of that character). For example, for a character "fins," the possible states may be "present" and "absent." For the character "number of forelimb digits," possible states may be 1, 2, 3, 4, or 5 3. For each character, determine which state is ancestral (primitive or plesiomorphic) and which is derived (apomorphic). This is usually done by comparison with a more distantly related organism termed the "outgroup." It is hypothesized that traits shared with the more distantly related organism(s) are likely to be "ancient" or plesiomorphic traits. Similarly, traits that differ from the outgroup are postulated to have arisen since the group being considered branched from its shared common ancestor with the outgroup, and thus are likely to be "derived" or apomorphic. 4. Group taxa by shared derived character states (synapomorphines). 5. When in doubt, choose the most parsimonious tree. While similar structures may evolve independently in separate lineages facing similar selective pressures (convergent evolution), this is assumed to be a rare event. Most major structures (eyes, horns, tails, fur, etc.) are assumed to have evolved or to be lost only rarely. Thus, when in doubt, choose a pathway that minimizes the number of times a feature must be postulated to have arisen (or lost) separately. An Example of Cladogram Construction for Vertebrates Trait Outgroup Frog (Lobefinned fish) Turtle Kangaroo Mouse Human Dorsal Nerve Yes Cord Yes Yes Yes Yes Yes Legs No Yes Yes Yes Yes Yes Nature of egg Requires water Requires Hard shell water prevents drying Develops inside the mother Develops inside the mother Develops inside the mother Nature of In egg development In egg In egg Marsupial Placental Placental Hair No No No Yes Yes Reduced Presence of pouch No No No Yes No No Bidpedal posture No No No Yes No Yes In this example, frogs share all major traits with the outgroup (i.e., they show mostly ancestral or plesiomorphic traits), except that they have legs and slightly enlarged brains. These last two features are apomorphies that are widespread in the vertebrate lineage. Frogs are thus postulated to have branched from the main vertebrate lineage relatively early in the evolutionary process. Turtles show further modifications from the outgroup, most markedly the presence of a hard shelled egg, as well as an increased tendency toward larger brain size; therefore we would suggest that their lineage branched next from the ancestral lineage. All three of the remaining groups are characterized by an egg that develops inside the mother, suggesting that these three share a common ancestor not shared by frogs and turtles. Mice and kangaroos share similar hair amounts, while humans and kangaroos share a generally bipedal posture. So how do we know how to group these three organisms? Firstly, we would suspect that the possession of hair, even in reduced amounts, might link humans to kangaroos and mice. Secondly, we would look to the other traits possessed by these groups. Both mice and humans show placental development and thus lack a pouch. Thus we would tend to link these two groups together more closely and the kangaroo more distantly. We would thus conclude that the cladogram for this group of organisms (minus the outgroup, which is not usually shown in these figures) should look something like the one below. You will note that, in this solution, the tendency toward a bipedal posture was postulated to have evolved twice; once in the marsupial lineage (kangaroos) and once in the placental lineage (humans). Such features are said to be analogous and to have resulted from convergent evolution. However, it is possible to draw the cladogram such that humans and kangaroos are postulated to have a common bipedal ancestor not shared by mice. However, in this solution, the tendency towards placental development, along with all the required anatomical changes (absence of a pouch etc.) must be postulated to have occurred twice (once in the mouse lineage and once in the human lineage). Such a solution would require more evolutionary steps than the sequence that we have proposed here and, consequently, would not be as efficient or "parsimonious" as our initial solution. Exercises Click here for Exercise 1. Click here for Exercise 2. Click here for Exercise 3. Click here for Exercise 4. Evolutionary (Synthetic) Systematics Like cladistics, evolutionary systematics is based strongly on evolutionary relationships. However, its supporters suggest that the degree of genetic differences between lineages should be used in addition to their genealogical (evolutionary) similarities when developing taxonomic classifications. Among other ways in which evolutionary systematics and a cladistic analyses differ: evolutionary taxonomists are more tolerant of multiple branches developing synchronously from an ancestral line (rather then just two as in cladistics), and they tend to treat organisms that are unchanged when a new branch arises from their lineage as the same species that existed before the branching event (i.e., the original species is not always believed to become extinct every time a branching event occurs). However, the biggest difference between the two approaches is that, when creating groupings, evolutionary taxonomists seek to maximize the way in which the groupings they create communicate information about the group of organisms in that taxon (in the way the word "mammal" or "reptile" does for even an untrained person). This type of division is also intended to increase the ease with which a field biologist, or even someone coming in from outside, can retrieve information about a group of organisms of interest to him/her using the taxonomic divisions formed. Taxa generated by an evolutionary biologist will never be polyphyletic, but may be either monophyletic or paraphyletic in nature. For example, as we saw earlier, birds and crocodilians diverged from the same ancestral reptilian line. A cladist would insist that these "sister groups" be placed in the same taxon, even though the amount of change from the common ancestor is much greater for the birds than it is for the crocodiles. An evolutionary taxonomist would suggest that the large number of similarities between crocodilians and reptiles would justify grouping them within the same general taxon, while placing birds in a separate taxon due to the large number of unique characters possessed by members of this group.