Word file (35 KB )
advertisement

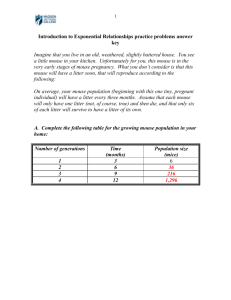
Table SI 4. Germline mouse mutations caused by viral retrotransposon insertion. Locus Adcybrl Mutant mouse name Strain of origin Early transposon (ETn) barreless ICR Cacng2stg stargazer Cacng2stg3J stargazer-3J Clc1adr myotonia Tnfrfs6lpr lymphoproliferative response EdarDl-slk sleek Fbxw4Dac dactylaplasia Fbxw4Dac-2J dactylaplasia-2J Fignfi fidget Foxn1nu-BC nude-British Columbia Gli3pdn extra-toes Hk1dea downeast anemia H2Q8 Reference 1 A/J 2 BALB/cJ 3 A2G 4 MRL/MpJ 5 STOCK Rb(8.12)5Bnr Tyrp1b m 6 SM/Ckc 7 MRL/MpJ 7 unknown/mixed 8 STOCK Spna1sph-2Bc 9 Jcl:ICR 10 A/J 11 MHC expression variant strain variant 12 Mip major intrinsic protein of eye lens fiber strain variant 13 MipCat-Fr Dominant cataract (Fraser) Mutedmu muted STOCK t 14 Lepob2J obese SM/Ckc-Fbxw4Dac 15 Tyrc-BC3 albino-British Columbia 3 STOCK Spna1sph-2Bc 9 vy A/J Intracisternal A-type Particle (IAP) C3H/HeJ A viable yellow Ahvy hypervariable yellow Aiapy agouti IAP yellow Atrnmg mahogany Atrnmg-L mahogany-Leicester AxinFu AxinKb 16 C3H/HeSnJ 17 B6C3F1/J 18 C3H x Swiss stock 19 C3H/HeJ 19 fused unspecified 20 knobbly unspecified 20 Dab1scm scrambler Dc/Le 21 Eya1bor branchio otorenal syndrome C3H/HeJ 22 GusMps2J mucopolysaccharidosis-3J C3H/HeOuJ 23 Hps1ep pale ear C3HeB/FeJ 24 Hps3coa6J cocoa-6J C3H/HeJ 25 Lamb3Iap Herlitz junctional epidermolysis bullosa C3H 26 Pitp1bvb vibrator DBA/2J 27 RelnAlb2J reelin-Albany-2 C3H/HeJ 28 CcS3/Dem 29 (101/Rl x C3H/Rl)F1 30 CBA/J 31 Spna1sph-Dem spherocytosis-Demant Tyrc-1R albino-1R Usp14ax ataxia Abcb1a Murine leukemia virus (MLV) multiple drug resistance-3 strain variant AifHq harlequin Myo5ad dilute hr hairless Pde6brd1 retinal-degeneration 1 32 CF1 33 fancy mice 34 unknown 35 fancy mice 36 fancy mice 37 Virus-like 30 (VL30) a 1 2 3 4 5 6 7 non-agouti Leong, W. L. et al. ETn insertion in the mouse Adcy1 gene: transcriptional and phylogenetic analyses. Mamm Genome 11, 97-103 (2000). Letts, V. A. et al. The mouse stargazer gene encodes a neuronal Ca2+ channel g subunit. Nat Genet 19, 340-347 (1998). Frankel, W. N. & Letts, V. A. (unpublished results). Schnulle, V. et al. The mouse Clc1/myotonia gene: ETn insertion, a variable AATC repeat, and PCR diagnosis of alleles. Mamm Genome 8, 718-725 (1997). Adachi, M., Watanabe-Fukunaga, R. & Nagata, S. Aberrant transcription caused by the insertion of an early transposable element in an intron of the Fas antigen gene of lpr mice. Proc Natl Acad Sci USA 90, 1756-1760 (1993). Headon, D. J. & Overbeek, P. A. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat Genet 22, 370374 (1999). Sidow, A. et al. A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet 23, 104-107 (1999). 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 Cox, G. A., Mahaffey, C. L., Nystuen, A., Letts, V. A. & Frankel, W. N. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nature Genet 26, 198-202 (2000). Hofmann, M., Harris, M., Juriloff, D. & Boehm, T. Spontaneous mutations in SELH/Bc mice due to insertions of early transposons: molecular characterization of null alleles at the nude and albino loci. Genomics 52, 107-109 (1998). Thien, H. & Ruther, U. The mouse mutation Pdn (Polydactyly Nagoya) is caused by the integration of a retrotransposon into the Gli3 gene. Mamm Genome 10, 205-209 (1999). Peters, L. L. et al. Downeast anemia (dea), a new mosue model of severe nonspherocytic hemolytic anemia caused by hexokinase (HK(I)) deficiency. Blood Cells Mol Dis 27, 850-860 (2001). Elliott, E., Rathbun, D., Ramsingh, A., Garberi, J. & Flaherty, L. Genetics and expression of the Q6 and Q8 genes. An LTR-like sequence in the 3' untranslated region. Immunogenetics 29, 371379 (1989). Shiels, A., Mackay, D., Bassnett, S., Al-Ghoul, K. & Kuszak, J. Disruption of lens fiber cell architecture in mice expressing a chimeric AQP0-LTR protein. Faseb J 14, 2207-2212 (2000). Zhang, Q. et al. The gene for the muted (mu) mouse, a model for Hermansky-Pudlak syndrome, defines a novel protein which regulates vesicle trafficking. Hum Mol Genet 11, 697-706 (2002). Moon, B. C. & Friedman, J. M. The molecular basis of the obese mutation in ob2J mice. Genomics 42, 152-156 (1997). Morgan, H. D., Sutherland, H. G., Martin, D. I. & Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23, 314-318 (1999). Argeson, A. C., Nelson, K. K. & Siracusa, L. D. Molecular basis of the pleiotropic phenotype of mice carrying the hypervariable yellow (Ahvy) mutation at the agouti locus. Genetics 142, 557-567 (1996). Michaud, E. J. et al. A molecular model for the genetic and phenotypic characteristics of the mouse lethal yellow (Ay) mutation. Proc Natl Acad Sci USA 91, 2562-2566 (1994). Gunn, T. M. et al. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature 398, 152-156 (1999). Vasicek, T. J. et al. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics 147, 777786 (1997). Sheldon, M. et al. Scrambler and yotari disrupt the disabled gene and produce a reeler- like phenotype in mice. Nature 389, 730-733 (1997). Johnson, K. R. et al. Inner ear and kidney anomalies caused by IAP insertion in an intron of the Eya1 gene in a mouse model of BOR syndrome. Hum Mol Genet 8, 645-653 (2000). Gwynn, B., Lueders, K., Sands, M. S. & Birkenmeier, E. H. Intracisternal A-particle element transposition into the murine beta-glucuronidase gene correlates with loss of enzyme activity: a new model for beta-glucuronidase deficiency in the C3H mouse. Mol Cell Biol 18, 6474-6481 (1998). Gardner, J. M. et al. The mouse pale ear (ep) mutation is the homologue of human Hermansky-Pudlak syndrome. Prot Natl Acad Sci U S A 94, 9238-9243 (1997). 25 26 27 28 29 30 31 32 33 34 35 36 37 Suzuki, T. et al. The gene mutated in cocoa mice, carrying a defect of organelle biogenesis, is a homologue of the human Hermansky-Pudlak syndrome-3 gene. Genomics 78, 30-37 (2001). Kuster, J. E., Guarnieri, M. H., Ault, J. G., Flaherty, L. & Swietek, P. J. IAP insertion in the murine LamB3 gene results in junctional epidermolysis bullosa. Mamm Genome 8, 673-681 (1997). Hamilton, B. A. et al. The vibrator mutation causes neurodegeneration via reduced expression of PITP alpha: positional complementation cloning and extragenic suppression. Neuron 18, 711-722 (1997). Royaux, I., Bernier, B., Montgomery, J. C., Flaherty, L. & Goffinet, A. M. Reln(rl-Alb2), an allele of reeler isolated from a chlorambucil screen, is due to an IAP insertion with exon skipping. Genomics 42, 479-482 (1997). Wandersee, N. J. et al. Defective spectrin integrity and neonatal thrombosis in the first mouse model for severe hereditary elliptocytosis. Blood 97, 543-550 (2001). Wu, M., Rinchik, E. M., Wilkinson, E. & Johnson, D. K. Inherited somatic mosaicism caused by an intracisternal A particle insertion in the mouse tyrosinase gene. Proc Natl Acad Sci U S A 94, 890-494 (1997). Wilson, S. M. et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet 10, in press (2002). Jun, K., Lee, S. B. & Shin, H. S. Insertion of a retroviral solo long terminal repeat in mdr-3 locus disrupts mRNA splicing in mice. Mamm Genome 11, 843-848 (2000). Klein, J. A. et al. The harlequin mouse mutation down-regulates apoptosis-inducing factor. Nature in press (2002). Jenkins, N. A., Copeland, N. G., Taylor, B. A. & Lee, B. K. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature 293, 370-374 (1981). Stoye, J. P., Fenner, S., Greenoak, G. E., Moran, C. & Coffin, J. M. Role of endogenous proviruses as insertional mutagens: The hairless mutation of mice. Cell 54, 383-391 (1988). Bowes, C. et al. Localization of a retroviral element within the gene coding for the b-subunit of cGMP-phosphodiesterase. Proc. Natl. Acad. Sci, USA 90, 2955-2959 (1993). Bultman, S. J. et al. Molecular analysis of reverse mutations from nonagouti (a) to black and tan (at) and white-bellied agouti (Aw) reveals alternative forms of agouti transcripts. Genes Dev 8, 481-490 (1994).