Downloaded
advertisement
Online supporting information
to accompany Lengyel et al. “Ants sow the seeds of global diversification in flowering plants”
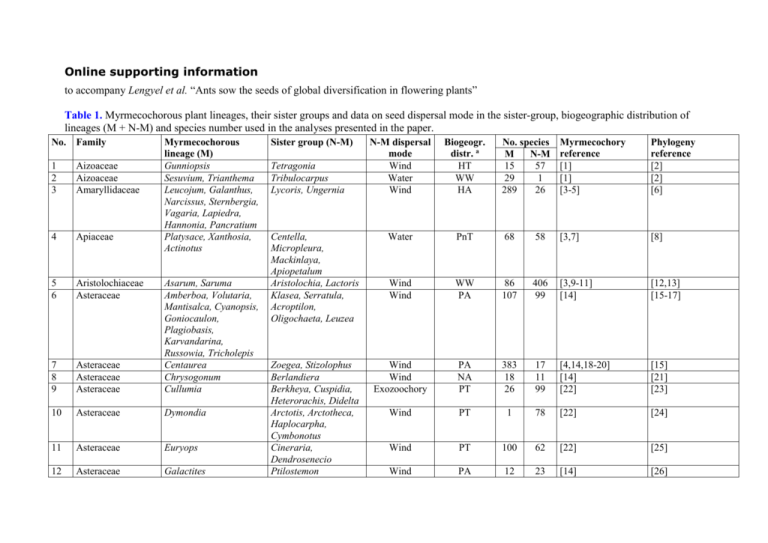
Table 1. Myrmecochorous plant lineages, their sister groups and data on seed dispersal mode in the sister-group, biogeographic distribution of
lineages (M + N-M) and species number used in the analyses presented in the paper.
No. Family
Myrmecochorous
lineage (M)
Gunniopsis
Sesuvium, Trianthema
Leucojum, Galanthus,
Narcissus, Sternbergia,
Vagaria, Lapiedra,
Hannonia, Pancratium
Platysace, Xanthosia,
Actinotus
1
2
3
Aizoaceae
Aizoaceae
Amaryllidaceae
4
Apiaceae
5
6
Aristolochiaceae
Asteraceae
7
8
9
Asteraceae
Asteraceae
Asteraceae
Asarum, Saruma
Amberboa, Volutaria,
Mantisalca, Cyanopsis,
Goniocaulon,
Plagiobasis,
Karvandarina,
Russowia, Tricholepis
Centaurea
Chrysogonum
Cullumia
10
Asteraceae
Dymondia
11
Asteraceae
Euryops
12
Asteraceae
Galactites
Sister group (N-M)
N-M dispersal
mode
Wind
Water
Wind
Biogeogr.
distr. a
HT
WW
HA
Centella,
Micropleura,
Mackinlaya,
Apiopetalum
Aristolochia, Lactoris
Klasea, Serratula,
Acroptilon,
Oligochaeta, Leuzea
Water
Zoegea, Stizolophus
Berlandiera
Berkheya, Cuspidia,
Heterorachis, Didelta
Arctotis, Arctotheca,
Haplocarpha,
Cymbonotus
Cineraria,
Dendrosenecio
Ptilostemon
Tetragonia
Tribulocarpus
Lycoris, Ungernia
No. species
M
N-M
15
57
29
1
289
26
Myrmecochory
reference
[1]
[1]
[3-5]
Phylogeny
reference
[2]
[2]
[6]
PnT
68
58
[3,7]
[8]
Wind
Wind
WW
PA
86
107
406
99
[3,9-11]
[14]
[12,13]
[15-17]
Wind
Wind
Exozoochory
PA
NA
PT
383
18
26
17
11
99
[4,14,18-20]
[14]
[22]
[15]
[21]
[23]
Wind
PT
1
78
[22]
[24]
Wind
PT
100
62
[22]
[25]
Wind
PA
12
23
[14]
[26]
No. Family
Myrmecochorous
lineage (M)
Osmitopsis
Osteospermum
Wedelia
Epimedium,
Vancouveria, Bongardia
13
14
15
16
Asteraceae
Asteraceae
Asteraceae
Berberidaceae
17
18
19
20
Berberidaceae
Boraginaceae
Bruniaceae
Bruniaceae
Gymnospermium
Nemophila
Audouinia capitata
Lonchostoma
21
Buxaceae
Buxus, Notobuxus
22
Cactaceae
Aztekium
23
Cactaceae
Gymnocalycium
24
25
Caricaceae
Caryophyllaceae
Carica
Moehringia
Sister group (N-M)
Athanasia
Calendula
Elaphandra
Podophyllum,
Sinopodophyllum,
Diphylleia, Dysosma,
Achlys
Leontice
Pholistoma
Tittmannia
Mniothamnea,
Raspalia
Sarcococca,
Pachysandra,
Styloceras
Echinocactus,
Astrophytum
Browningia, Cereus,
Coleocephalocereus,
Micranthocereus,
Samaipaticereus,
Echinopsis, Harrisia,
Haageocereus,
Matucana,
Oreocereus,
Rauhocereus
Moringa
Arenaria subg.
Arenaria, subg.
Leiosperma,
‘Pseudomoehringia’
group
N-M dispersal
mode
Wind
Wind
Passive
Endozoochory
Biogeogr.
distr. a
PT
OW
NW
HA
No. species
M
N-M
11
54
153
20
100
16
67
21
Myrmecochory
reference
[22]
[22]
[22,28]
[14,30-32]
Phylogeny
reference
[27]
[27]
[29]
[33-35]
Endozoochory
Water
Passive
Passive
HA
NA
PT
PT
12
11
1
10
28
3
25
4
[14]
[4,14,30,36]
[22]
[22]
[33-35]
[37]
[38]
[38]
Endozoochory
WW
90
20
[14,39]
[40]
Exozoochory
NW
2
10
[41]
[42]
Exozoochory
NW
70
1100 [41]
[42]
Wind
Wind
PnT
HA
23
31
12
172
[43]
[45]
[14]
[4,14,44]
No. Family
26
Colchicaceae
27
28
Dasypogonaceae
Dilleniaceae
29
30
31
32
Dipsacaceae
Dipsacaceae
Elaeocarpaceae
Euphorbiaceae
33
Euphorbiaceae
34
35
Euphorbiaceae
Euphorbiaceae
Myrmecochorous
lineage (M)
Colchicum,
Androcymbium,
Hexacyrtis,
Ornithoglossum,
Sandersonia, Gloriosa,
Baeometra, Wurmbea,
Camptorrhiza,
Iphigenia,
Schelhammera,
Tripladenia, Disporum,
Uvularia, Kuntheria,
Dasypogon
Hibbertia, Adrastaea,
Pachynema
Knautia
Scabiosa
Tetratheca
Astraea
Bertya, Beyeria,
Ricinocarpos
Claoxylon
Conceveiba
Sister group (N-M)
Burchardia
N-M dispersal
mode
Passive
Biogeogr.
distr. a
WW
No. species Myrmecochory
M
N-M reference
245
5
[3,7,10,46-48]
Phylogeny
reference
[49]
Calectasia
Dillenia
Passive
Endozoochory
AU
PnT
4
124
11
60
[47]
[3,7]
[50]
[51]
Pterocephalidium
Sixalix
Platytheca
Ophellantha,
Acidocroton
Baloghia, Fontainea
Wind
Wind
Water
Endozoochory
PA
PA
AU
NT
60
80
39
10
1
10
2
12
[10,32,52]
[14]
[3,7,54,55]
[57]
[53]
[53]
[54,56]
[57]
Endozoochory
AU
45
29
[3,7,14]
[58]
Erythrococca
Endozoochory
Gavarretia, Polyandra
Passive
OW
NT
75
18
50
2
[7]
[60]
[58,59]
[58]
No. Family
Myrmecochorous
lineage (M)
Euphorbia,
Chamaesyce,
Synadenium,
Monadenium,
Pedilanthus,
Neoguillauminia,
Calycopeplus,
Anthosterna,
Dichostemma
36
Euphorbiaceae
37
Euphorbiaceae
Monotaxis, Adriana,
Amperea
38
Euphorbiaceae
39
40
Euphorbiaceae
Fabaceae
Pera, Clutia,
Chaetocarpus
Seidelia
Cytisus
Sister group (N-M)
Spirostachys,
Excoecaria,
Sebastiana,
Colliguaja, Sapium,
Stillingia, Adenopeltis,
Grimmeodendron,
Bonania, Hippomane,
Ophthalmoblapton,
Tetraplandra,
Pachystroma, Hura,
Gymnanthes, Mabea,
Microstachys,
Homalanthus,
Neoshirakia,
Dalembertia,
Triadica,
Actinostemon,
Pseudosenefeldera,
Senefelderopsis,
Maprounea
Cephalomappa,
Koilodepas,
Cephalocroton,
Sumbaviopsis,
Melanolepis,
Chrozophora,
Discocleidion,
Ricinus, Speranskia
Pogonophora
Leidesia
Cytisophyllum,
Laburnum
N-M dispersal
mode
Endozoochory
Biogeogr.
distr. a
WW
No. species Myrmecochory
M
N-M reference
1846 407 [3,4,61,62]
Phylogeny
reference
[58]
Passive
OW
31
37
[3,60]
[58]
Wind
PnT
108
6
[22,58,60]
[58]
Passive
Wind
PT
PA
4
60
1
3
[60]
[63,64]
[58]
[65,66]
No. Family
41
Fabaceae
42
Fabaceae
43
Fabaceae
44
Fabaceae
45
Fabaceae
46
47
48
49
Gesneriaceae
Gesneriaceae
Goodeniaceae
Goodeniaceae
50
Gyrostemonaceae
51
Hemerocallidaceae
52
53
54
Hyacinthaceae
Hyacinthaceae
Iridaceae
Myrmecochorous
lineage (M)
Daviesia, Viminaria,
Erichsenia, Goodia,
Bossiaea, Platylobium,
Muelleranthus,
Ptychosema,
Aenictophyton
Hardenbergia,
Kennedia
Sister group (N-M)
Gompholobium,
Sphaerolobium
Mucuna, Desmodium,
Lespedeza,
Kummerowia,
Campylotropis
Hovea, Templetonia,
Brongniartia,
Lamprolobium
Plagiocarpus
Pultenaea (= Mirbelia s. Isotropis
l.)
Ulex, Stauracanthus
Genista sect.
Cephalospartum
Chrysothemis
Nautilocalyx
Codonanthe
Nematanthus
Dampiera
Anthotium
Goodenia, Scaevola,
Brunonia
Verreauxia, Velleia,
Coopernookia
Gyrostemon,
Tovaria
Codonocarpus,
Walteranthus, Tersonia,
Reseda
Caesia, Hensmania,
Tricoryne
Johnsonia, Arnocrinum
Lachenalia
Polyxena, Periboea
Scilla, Chionodoxa
Bellevalia
Witsenia, Klattia
Nivenia
N-M dispersal
mode
Passive
Biogeogr.
distr. a
AU
No. species Myrmecochory
M
N-M reference
287
43 [3,7,67]
Phylogeny
reference
[68]
Water
PnT
59
500
[3,69,70]
[71]
Wind
HT
52
57
[3,7]
[72]
Passive
AU
470
16
[3,7,70,73]
[68,74]
Passive
HA
21
5
[4,18,52]
[66]
Endozoochory
Endozoochory
Wind
Passive
NT
NT
AU
AU
12
34
66
350
55
48
5
3
[32,75]
[14]
[3,7]
[3,7,69]
[76]
[76]
[77]
[77]
Passive
AU
18
2
[3]
[43,78]
Exozoochory
AU
58
12
[3,5,7]
[50]
Passive
Wind
Passive
PT
PA
PT
110
40
4
23
50
10
[14]
[4]
[81]
[79]
[80]
[82,83]
No. Family
55
Juncaceae
56
Lamiaceae
57
58
59
Lamiaceae
Lamiaceae
Lamiaceae
60
Liliaceae
61
62
63
64
Liliaceae
Liliaceae
Limeaceae
Malvaceae
65
Malvaceae
66
Melanthiaceae
67
Papaveraceae
Myrmecochorous
lineage (M)
Luzula
Sister group (N-M)
Juncus, Distichia,
Marsippospermum,
Rostkovia
Ajuga
Caryopteris,
Trichostema
Lamium
Marrubium
Rosmarinus
Perovskia
Teucrium
Teucridium,
Oncinocalyx
Erythronium spp. Erythronium spp. Eurasian + Eastern
Western North
North American lineage American lineage
Gagea
Lloydia
Scoliopus
Streptopus
Macarthuria
Limeum
Gossypium sect.
Gossypium sect.
Grandicalyx ('genome
Sturtia ('genom C'),
K' lineage)
sect. Hibiscoidea
('genome G')
Lasiopetalum,
Hermannia,
Hannafordia,
Waltheria, Melochia
Maxwellia, Thomasia,
Guichenotia,
Commersonia, Rulingia,
Keraudrenia, Seringia
Trillium
Paris, Daiswa,
Kinugasa, Trillidium
Corydalis, Dicentra,
Hypecoum
Adlumia,
Dactylocapnos,
Rupicapnos,
Pseudofumaria,
Cysticapnos
N-M dispersal
mode
Water
Biogeogr.
distr. a
WW
No. species Myrmecochory
M
N-M reference
115
249 [3,9,20]
Phylogeny
reference
[84]
Passive
WW
50
37
[3,85]
[86]
Exozoochory
Wind
Exozoochory
HA
PA
WW
190
22
250
139
10
2
[4,14]
[14,85]
[14]
[86]
[87]
[86]
Passive
HA
8
17
[5,88,89]
[90]
Passive
Endozoochory
Endozoochory
Water
HA
HA
OW
AU
91
5
12
12
12
30
21
5
[5,88]
[14,88,92]
[3,73]
[3,7]
[50,88,91]
[88]
[93]
[94,95]
Endozoochory
WW
167
205
[3,7,70]
[96]
Endozoochory
HA
38
27
[5,9,14]
[97]
Exozoochory
HA
384
18
[9,22]
[98]
No. Family
68
Papaveraceae
69
Penaeaceae
70
71
Phyllanthaceae
Picrodendraceae
72
73
74
Poaceae
Poaceae
Polygalaceae
75
76
77
Portulacaceae
Primulaceae
Proteaceae
Myrmecochorous
lineage (M)
Sanguinaria,
Chelidonium, Eomecon,
Macleaya, Bocconia,
Hylomecon,
Stylophorum
Penaea, Brachysiphon,
Endonema, Saltera,
Stylapterus,
Sonderothamnus
Breynia
Picrodendron,
Micrantheum,
Oldfieldia,
Stachystemon,
Aristogeitonia, Scagea,
Tetracoccus
Chionachne
Melica
Polygala, Bredemeyera,
Muraltia, Nylandtia,
Heterosamara,
Salomonia,
Comesperma, Monnina,
Securidaca
Claytonia, Montia
Cyclamen
Grevillea
Sister group (N-M)
Dicranostigma
N-M dispersal
mode
Passive
Biogeogr.
distr. a
HA
No. species Myrmecochory
M
N-M reference
99
8
[4,9,10,14]
Phylogeny
reference
[99]
Olinia
Endozoochory
PT
23
8
[22]
[100-102]
Passive
Passive
IM
WW
35
82
40
3
[3,7]
[3,7,58,62,104]
[103]
[105], Wurdack
unpubl. from [58]
Phacelurus
Glyceria
Carpolobia, Atroxima
Wind
Water
Endozoochory
IM
HA
WW
9
80
910
10
35
6
[32]
[4,14,30]
[3,7,14,108]
[106]
[107]
[108]
Lewisia
Coris
Buckinghamia,
Opisthiolepis
Wind
Exozoochory
Wind
NW
PA
AU
41
20
260
16
3
3
[9,14,32]
[14,32]
[3,7,112]
[109,110]
[111]
[113,114]
Sauropus
Podocalyx,
Paradrypetes
No. Family
78
Proteaceae
79
80
81
82
Ranunculaceae
Ranunculaceae
Ranunculaceae
Ranunculaceae
83
84
Ranunculaceae
Restionaceae
85
86
Rhamnaceae
Rhamnaceae
87
Rubiaceae
88
Rutaceae
89
Rutaceae
90
91
Santalaceae
Sapindaceae
Myrmecochorous
lineage (M)
Mimetes, Orothamnus,
Leucospermum,
Diastella, Sorocephalus,
Spatalla, Paranomus,
Vexatorella, Serruria,
Leucadendron,
Adenanthos
Anemone
Delphinium
Ficaria
Helleborus
Sister group (N-M)
Isopogon
N-M dispersal
mode
Wind
Pulsatilla, Knowltonia
Wind
Consolida
Wind
Coptidium
Passive
Myosurus, Nigella,
Passive
Actaea, Cimicifuga
Trollius, Adonis
Caltha
Water
Restio
Elegia,
Wind
Chondropetalum,
Dovea
Phylica, Trichocephalus Nesiota, Noltea
Endozoochory
Pomaderris, Spyridium, Colletia, Discaria,
Passive
Trymalium, Siegfriedia, Alphitonia, Granitites
Cryptandra,
Stenanthemum
Opercularia, Pomax
Leptostigma,
Endozoochory
Durlingtonia, Nertera,
Normandia,
Coprosma
Asterolasia
Drummondita,
Endozoochory
Muiriantha
Phebalium, Microcybe Nematolepis,
Passive
Rhadinothamnus,
Chorilaena
Thesium, Osyridicarpos Buckleya
Endozoochory
Cardiospermum
Paullinia
Endozoochory
Biogeogr.
distr. a
HT
No. species Myrmecochory
M
N-M reference
308
65 [3,115]
Phylogeny
reference
[113,114,116]
HA
HA
PA
HA
120
250
5
20
48
40
2
54
[4,9,14]
[14]
[118]
[4,14,32,120]
[117]
[117]
[119]
[117]
HA
PT
55
88
10
73
[14]
[3,7]
[117]
[84,121]
PT
HT
151
172
2
50
[22]
[3,7,54,69]
[122]
[122]
HT
19
112
[7]
[123,124]
AU
21
9
[125]
[126]
AU
31
11
[3,7,125]
[126]
WW
PnT
177
61
7
195
[52,127]
[129]
[128]
[130]
No. Family
92
93
Sapindaceae
Scrophulariaceae
Myrmecochorous
lineage (M)
Dodonaea
Melampyrum
94
Scrophulariaceae
Pedicularis
95
96
97
98
99
100
101
Solanaceae
Tecophilaeaceae
Turneraceae
Urticaceae
Valerianaceae
Violaceae
Zygophyllaceae
Datura
Cyanastrum
Turnera
Parietaria
Fedia
Viola
Zygophyllum, Augea,
Fagonia
a
Sister group (N-M)
Distichostemon
Bartsia, Lathraea,
Rhinanthus,
Rhynchocorys,
Parentucellia,
Odontites, Euphrasia,
Tozzia
Agalinis, Esterhazya,
Aureolaria, Seymeria,
Lamourouxia,
Castilleja,
Orthocarpus,
Cordylanthus,
Triphysaria
Brugmansia
Walleria
Piriqueta
Boehmeria
Valerianella
Noisettia
Larrea, Pintoa,
Bulnesia, Porlieria,
Guaiacum
N-M dispersal
mode
Passive
Wind
Biogeogr.
distr. a
HT
HA
No. species
M
N-M
70
8
35
310
Myrmecochory
reference
[3,131,132]
[4,14,30,118,133]
Phylogeny
reference
[130]
[134,135]
Wind
HA
350
341
[4,14,30,133]
[134,135]
Passive
Passive
Passive
Exozoochory
Wind
Passive
Wind
NW
PT
NT
WW
PA
WW
WW
11
7
100
10
3
400
120
5
3
44
80
50
8
31
[14]
[14]
[137,138]
[10,32]
[14]
[3,4,7,9,10,20]
[22]
[136]
[82]
[139]
[140]
[141]
[142]
[143,144]
Biogeographic distribution types for lineages comprising both M and N-M lineages:
AU – Australian, IM – Indo-Malayan, NA – Nearctic, NT – Neotropical, PA – Palearctic, PT – Paleotropical (mostly Cape Floristic Region);
complex distribution types: HA – Holarctic (NA + PA), NW – New World (NA + NT), OW – Old World (AU + IM + PA + PT), PnT – Pantropical (AU + IM + NT + PT), HT – Holotropical (AU + NT + PT), WW – Worldwide.
References
1. Hassan NMS, Meve U, Liede-Schumann S (2005) Seed coat morphology of Aizoaceae-Sesuvioideae, Gisekiaceae and Molluginaceae and its
systematic significance. Botanical Journal of the Linnean Society 148: 189-206.
2. Klak C, Khunou A, Reeves G, Hedderson T (2003) A phylogenetic hypothesis for the Aizoaceae (Caryophyllales) based on four plastid DNA
regions. American Journal of Botany 90: 1433-1445.
3. Berg RY (1975) Myrmecochorous plants in Australia and their dispersal by ants. Australian Journal of Botany 23: 475-508.
4. van der Pijl L (1982) Principles of Dispersal in Higher Plants. Berlin: Springer-Verlag. 215 p.
5. Dahlgren RMT, Clifford HT, Yeo PF (1985) The Families of the Monocotyledons: Structure, Evolution, and Taxonomy. Berlin: SpringerVerlag. 520 p.
6. Meerow AW, Francisco-Ortega J, Kuhn DN, Schnell RJ (2006) Phylogenetic relationships and biogeography within the Eurasian clade of
Amaryllidaceae based on plastid ndhF and nrDNA ITS sequences: Lineage sorting in a reticulate area? Systematic Botany 31: 42-60.
7. Westoby M, Rice B, Howell J (1990) Seed size and plant growth form as factors in dispersal spectra. Ecology 71: 1307-1315.
8. Chandler GT, Plunkett GM (2004) Evolution in Apiales: nuclear and chloroplast markers together in (almost) perfect harmony. Botanical
Journal of the Linnean Society 144: 123-147.
9. Beattie AJ, Culver DC (1981) The guild of myrmecochores in the herbaceous flora of West Virginia forests. Ecology 62: 107-115.
10. Gorb E, Gorb S (2003) Seed Dispersal by Ants in a Deciduous Forest Ecosystem. Dordrecht: Kluwer Academic Publishers. 225 p.
11. Gonzalez F, Rudall PJ (2003) Structure and development of the ovule and seed in Aristolochiaceae, with particular reference to Saruma.
Plant Systematics and Evolution 241: 223-244.
12. Kelly LM (1998) Phylogenetic relationships in Asarum (Aristolochiaceae) based on morphology and ITS sequences. American Journal of
Botany 85: 1454-1467.
13. Neinhuis C, Wanke S, Hilu KW, Muller K, Borsch T (2005) Phylogeny of Aristolochiaceae based on parsimony, likelihood, and Bayesian
analyses of trnL-trnF sequences. Plant Systematics and Evolution 250: 7-26.
14. Beattie AJ (1983) Distribution of ant-dispersed plants. In: Kubitzki K, editor. Dispersal and distribution. Hamburg: Sonderbaende des
Naturwissenschaftlichen Vereins - Parey. pp. 249-270.
15. Garcia-Jacas N, Susanna A, Garnatje T, Vilatersana R (2001) Generic delimitation and phylogeny of the subtribe Centaureinae (Asteraceae):
A combined nuclear and chloroplast DNA analysis. Annals of Botany 87: 503-515.
16. Martins L, Hellwig FH (2005) Systematic position of the genera Serratula and Klasea within Centaureinae (Cardueae, Asteraceae) inferred
from ETS and ITS sequence data and new combinations in Klasea. Taxon 54: 632-638.
17. Petit DP (1997) Generic interrelationships of the Cardueae (Compositae): a cladistic analysis of morphological data. Plant Systematics and
Evolution 207: 173-203.
18. Melendo M, Gimenez E, Cano E, Gomez-Mercado F, Valle F (2003) The endemic flora in the south of the Iberian Peninsula: taxonomic
composition, biological spectrum, pollination, reproductive mode and dispersal. Flora 198: 260-276.
19. Wagenitz G, Hellwig FH (1996) Evolution of characters and phylogeny of the Centaureinae. In: N. HDJ, Beentje HJ, editors. Compositae:
Systematics Proceedings of the International Compositae Conference, Kew 1994, Vol 1. London: Kew: Royal Botanic Gardens. pp. 491510.
20. Andersen UV (1993) Dispersal strategies of Danish seashore plants. Ecography 16: 289-298.
21. Clevinger JA, Panero JL (2000) Phylogenetic analysis of Silphium and subtribe Engelmanniinae (Asteraceae: Heliantheae) based on ITS and
ETS sequence data. American Journal of Botany 87: 565-572.
22. Bond WJ, Slingsby P (1983) Seed dispersal by ants in shrublands of the Cape Province and its evolutionary implications. South African
Journal of Science 79: 231-233.
23. Karis PO (2006) Morphological data indicates two major clades of the subtribe Gorterlinae (Asteraceae-Arctotideae). Cladistics 22: 199-221.
24. McKenzie RJ, Muller EM, Skinner AKW, Karis PO, Barker NP (2006) Phylogenetic relationships and generic delimitation in subtribe
Arctotidinae (Asteraceae : Arctotideae) inferred by DNA sequence data from ITS and five chloroplast regions. American Journal of
Botany 93: 1222-1235.
25. Knox EB, Palmer JD (1995) Chloroplast DNA variation and the recent radiation of the siant Senecios (Asteraceae) on the tall mountains of
Eastern Africa. Proceedings of the National Academy of Sciences of the United States of America 92: 10349-10353.
26. Garcia-Jacas N, Garnatje T, Susanna A, Vilatersana R (2002) Tribal and subtribal delimitation and phylogeny of the Cardueae (Asteraceae):
A combined nuclear and chloroplast DNA analysis. Molecular Phylogenetics and Evolution 22: 51-64.
27. Watson LE, Evans TM, Boluarte T (2000) Molecular phylogeny and biogeography of tribe Anthemidae (Asteraceae), based on chloroplast
gene ndhF. Molecular Phylogenetics and Evolution 15: 59-69.
28. Nesom GL (1981) Ant dispersal in Wedelia hispida HBK. (Heliantheae: Compositae). Southwestern Naturalist 26: 5-12.
29. Panero JL, Jansen RK, Clevinger JA (1999) Phylogenetic relationships of subtribe Ecliptinae (Asteraceae : Heliantheae) based on chloroplast
DNA restriction site data. American Journal of Botany 86: 413-427.
30. Stebbins GL (1971) Adaptive radiation of reproductive characteristics in angiosperms, II: seeds and seedlings. Annual Review of Ecology
and Systematics 2: 237-260.
31. Berg RY (1972) Dispersal ecology of Vancouveria (Berberidaceae). American Journal of Botany 59: 109-&.
32. Mabberley DJ (2008) Mabberley's Plant Book. A portable dictionary of plants, their classification and uses. Cambridge: Cambridge
University Press. 1021 p.
33. Kim YD, Jansen RK (1998) Chloroplast DNA restriction site variation and phylogeny of the Berberidaceae. American Journal of Botany 85:
1766-1778.
34. Kim YD, Kim SH, Kim CH, Jansen RK (2004) Phylogeny of Berberidaceae based on sequences of the chloroplast gene ndhF. Biochemical
Systematics and Ecology 32: 291-301.
35. Liu JQ, Chen ZD, Lu AM (2002) Molecular evidence for the sister relationship of the eastern Asia-North American intercontinental species
pair in the Podophyllum group (Berberidaceae). Botanical Bulletin of Academia Sinica 43: 147-154.
36. Chuang TI, Constance L (1992) Seeds and systematics in Hydrophyllaceae - Tribe Hydrophylleae. American Journal of Botany 79: 257-264.
37. Ferguson DM (1998) Phylogenetic analysis and relationships in Hydrophyllaceae based on ndhF sequence data. Systematic Botany 23: 253268.
38. Quint M, Classen-Bockhoff R (2006) Phylogeny of Bruniaceae based on matK and its sequence data. International Journal of Plant Sciences
167: 135-146.
39. Kohler E (2007) Buxaceae. In: Kubitzki K, Bayer C, Stevens PF, editors. The Families and Genera of Vascular Plants Vol 9, Flowering
Plants Eudicots: Berberidopsidales, Buxales, Crossosomatales, Fabales pp, Geraniales, Gunnerales, Myrtales pp, Proteales, Saxifragales,
Vitales, Zygophyllales, Clusiaceae Alliance, Passifloraceae Alliance, Dilleniaceae, Huaceae. Berlin: Springer. pp. 40-47.
40. von Balthazar M, Endress PK, Qiu YL (2000) Phylogenetic relationships in Buxaceae based on nuclear internal transcribed spacers and
plastid ndhF sequences. International Journal of Plant Sciences 161: 785-792.
41. Van Rheede KvO, Rooyen MWv (1999) Desert Plants. Berlin: Springer. 242 p.
42. Nyffeler R (2002) Phylogenetic relationships in the cactus family (Cactaceae) based on evidence from trnK/matK and trnL-trnF sequences.
American Journal of Botany 89: 312-326.
43. Rodman JE, Karol KG, Price RA, Sytsma KJ (1996) Molecules, morphology, and Dahlgren's expanded order Capparales. Systematic Botany
21: 289-307.
44. Minuto L, Fior S, Roccotiello E, Casazza G (2006) Seed morphology in Moehringia L. and its taxonomic significance in comparative studies
within the Caryophyllaceae. Plant Systematics and Evolution 262: 189-208.
45. Fior S, Karis PO (2007) Phylogeny, evolution and systematics of Moehringia (Caryophyllaceae) as inferred from molecular and
morphological data: a case of homology reassessment. Cladistics 23: 362-372.
46. Membrives N, Pedrola-Monfort J, Caujape-Castells J (2003) Morphological seed studies of Southwest African Androcymbium
(Colchicaceae). Botanica Macaronesica 24: 87-106.
47. Dunn RR, Gove AD, Barraclough TG, Givnish TJ, Majer JD (2007) Convergent evolution of an ant-plant mutualism across plant families,
continents, and time. Evolutionary Ecology Research 9: 1349-1362.
48. Nordenstam N (1998) Colchicaceae. In: Kramer KU, Kubitzki K, Huber H, Green PS, editors. The Families and Genera of Vascular Plants
Vol 3, Flowering Plants Monocotyledons: Lilianae (except Orchidaceae). Berlin: Springer. pp. 295-333.
49. Vinnersten A, Reeves G (2003) Phylogenetic relationships within Colchicaceae. American Journal of Botany 90: 1455-1462.
50. Givnish TJ, Pires JC, Graham SW, McPherson MA, Prince LM, et al. (2005) Repeated evolution of net venation and fleshy fruits among
monocots in shaded habitats confirms a priori predictions: evidence from an ndhF phylogeny. Proceedings of the Royal Society BBiological Sciences 272: 1481-1490.
51. Horn JW (2006) Dilleniaceae. In: Kubitzki K, Bayer C, Stevens PF, editors. The Families and Genera of Vascular Plants Vol 9, Flowering
Plants Eudicots: Berberidopsidales, Buxales, Crossosomatales, Fabales pp, Geraniales, Gunnerales, Myrtales pp, Proteales, Saxifragales,
Vitales, Zygophyllales, Clusiaceae Alliance, Passifloraceae Alliance, Dilleniaceae, Huaceae. Berlin: Springer. pp. 132-154.
52. Sernander R (1906) Entwurf einer Monographie der europäischen Myrmekochoren. Kunglica Svenska Vetenskapsakademien Handlingar 41:
1-410.
53. Caputo P, Cozzolino S, Moretti A (2004) Molecular phylogenetics of Dipsacaceae reveals parallel trends in seed dispersal syndromes. Plant
Systematics and Evolution 246: 163-175.
54. Boesewinkel FD (1999) Ovules and seeds of Tremandraceae. Australian Journal of Botany 47: 769-781.
55. Harrington GN, Driver MA (1995) The effect of fire and ants on the seed-bank of a shrub in a semi-arid grassland. Australian Journal of
Ecology 20: 538-547.
56. Crayn DM, Rossetto M, Maynard DJ (2006) Molecular phylogeny and dating reveals an Oligo-Miocene radiation of dry-adapted shrubs
(former Tremandraceae) from rainforest tree progenitors (Elaeocarpaceae) in Australia. American Journal of Botany 93: 1328-1342.
57. Berry PE, Hipp AL, Wurdack KJ, Van Ee B, Riina R (2005) Molecular phylogenetics of the giant genus Croton and tribe Crotoneae
(Euphorbiaceae sensu stricto) using ITS and trnL-trnF DNA sequence data. American Journal of Botany 92: 1520-1534.
58. Wurdack KJ, Hoffmann P, Chase MW (2005) Molecular phylogenetic analysis of uniovulate Euphorbiaceae (Euphorbiaceae sensu stricto)
using plastid rbcL and trnL-F DNA sequences. American Journal of Botany 92: 1397-1420.
59. Kabouw P, Van Welzen PC, Baas P, Van Heuven BJ (2008) Styloid crystals in Claoxylon (Euphorbiaceae) and allies (Claoxylinae) with
notes on leaf anatomy. Botanical Journal of the Linnean Society 156: 445-457.
60. Tokuoka T, Tobe H (2003) Ovules and seeds in Acalyphoideae (Euphorbiaceae): Structure and systematic implications. Journal of Plant
Research 116: 355-380.
61. Tokuoka T, Tobe H (2002) Ovules and seeds in Euphorbioideae (Euphorbiaceae): Structure and systematic implications. Journal of Plant
Research 115: 361-374.
62. Webster GL (1994) Classification of the Euphorbiaceae. Annals of the Missouri Botanical Garden 81: 3-32.
63. Pemberton RW, Irving DW (1990) Elaiosomes on weed seeds and the potential for myrmecochory in naturalized plants. Weed Science 38:
615-619.
64. Cristofolini G, Troia A (2006) A reassessment of the sections of the genus Cytisus Desf. (Cytiseae, Leguminosae). Taxon 55: 733-746.
65. Cubas P, Pardo C, Tahiri H (2002) Molecular approach to the phylogeny and systematics of Cytisus (Leguminosae) and related genera based
on nucleotide sequences of nrDNA (ITS region) and cpDNA (trnL-trnF intergenic spacer). Plant Systematics and Evolution 233: 223-242.
66. Pardo C, Cubas P, Tahiri H (2004) Molecular phylogeny and systematics of Genista (Leguminosae) and related genera based on nucleotide
sequences of nrDNA (ITS region) and cpDNA (trnL-trnF intergenic spacer). Plant Systematics and Evolution 244: 93-119.
67. de Kok RPJ, West JG (2002) A revision of Pultenaea (Fabaceae) - 1. Species with ovaries glabrous and/or with tufted hairs. Australian
Systematic Botany 15: 81-113.
68. Orthia LA, Crisp MD, Cook LG, de Kok RPJ (2005) Bush peas: a rapid radiation with no support for monophyly of Pultenaea (Fabaceae :
Mirbelieae). Australian Systematic Botany 18: 133-147.
69. Milewski AV, Bond WJ (1982) Convergence of myrmecochory (dispersal and sowing of seeds by ants) in mediterranean Australia and South
Africa. In: Buckley RC, editor. Ant-plant Interactions in Australia. The Hague: Junk. pp. 85-94.
70. Hughes L, Westoby M (1992) Fate of seeds adapted for dispersal by ants in Australian sclerophyll vegetation. Ecology 73: 1285-1299.
71. Kajita T, Ohashi H, Tateishi Y, Bailey CD, Doyle JJ (2001) rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae,
and allies. Systematic Botany 26: 515-536.
72. Thompson IR, Ladiges PY, Ross JH (2001) Phylogenetic studies of the tribe Brongniartieae (Fabaceae) using nuclear DNA (ITS-1) and
morphological data. Systematic Botany 26: 557-570.
73. Steyn EMA, van Wyk AE, Smith GF (2002) Ovule, seed and seedling characters in Acharia (Achariaceae) with evidence of myrmecochory
in the family. South African Journal of Botany 68: 143-156.
74. Crisp M, Cook L, Steane D (2004) Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell
us about the evolution of diversity in present-day communities? Philosophical Transactions of the Royal Society of London Series BBiological Sciences 359: 1551-1571.
75. Lu KL, Mesler MR (1981) Ant dispersal of a Neotropical forest floor gesneriad. Biotropica 13: 159-160.
76. Zimmer EA, Roalson EH, Skog LE, Boggan JK, Idnurm A (2002) Phylogenetic relationships in the Gesnerioideae (Gesneriaceae) based on
nrDNA ITS and cpDNA trnL-F and trnE-T spacer region sequences. American Journal of Botany 89: 296-311.
77. Gustafsson MHG, Backlund A, Bremer B (1996) Phylogeny of the Asterales sensu late based on rbcL sequences with particular reference to
the Goodeniaceae. Plant Systematics and Evolution 199: 217-242.
78. Hufford L (1996) Developmental morphology of female flowers of Gyrostemon and Tersonia and floral evolution among Gyrostemonaceae.
American Journal of Botany 83: 1471-1487.
79. Pfosser M, Wetschnig W, Ungar S, Prenner G (2003) Phylogenetic relationships among genera of Massonieae (Hyacinthaceae) inferred from
plastid DNA and seed morphology. Journal of Plant Research 116: 115-132.
80. Pfosser M, Speta F (1999) Phylogenetics of Hyacinthaceae based on plastid DNA sequences. Annals of the Missouri Botanical Garden 86:
852-875.
81. Manning JC, Goldblatt P (1991) Systematic and phylogenetic significance of the seed coat in the shrubby African Iridaceae, Nivenia, Klattia
and Witsenia. Botanical Journal of the Linnean Society 107: 387-404.
82. Chase MW, De Bruijn AY, Cox AV, Reeves C, Rudall PJ, et al. (2000) Phylogenetics of Asphodelaceae (Asparagales): An analysis of plastid
rbcL and trnL-F DNA sequences. Annals of Botany 86: 935-951.
83. Davies TJ, Barraclough TG, Savolainen V, Chase MW (2004) Environmental causes for plant biodiversity gradients. Philosophical
Transactions of the Royal Society of London Series B-Biological Sciences 359: 1645-1656.
84. Bremer K (2002) Gondwanan evolution of the grass alliance of families (Poales). Evolution 56: 1374-1387.
85. Hsi-wen L, Hedge IC (1994) Lamiaceae. Flora of China, vol 17. pp. 50-299.
86. Wagstaff SJ, Hickerson L, Spangler R, Reeves PA, Olmstead RG (1998) Phylogeny in Labiatae s.l., inferred from cpDNA sequences. Plant
Systematics and Evolution 209: 265-274.
87. Walker JB, Sytsma KJ, Treutlein J, Wink M (2004) Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and
ecological specializations of Salvia and tribe Mentheae. American Journal of Botany 91: 1115-1125.
88. Patterson TB, Givnish TJ (2002) Phylogeny, concerted convergence, and phylogenetic niche conservatism in the core Liliales: Insights from
rbcL and ndhF sequence data. Evolution 56: 233-252.
89. Guitian P, Medrano M, Guitian J (2003) Seed dispersal in Erythronium dens-canis L. (Liliaceae): variation among habitats in a
myrmecochorous plant. Plant Ecology 169: 171-177.
90. Allen GA, Soltis DE, Soltis PS (2003) Phylogeny and biogeography of Erythronium (Liliaceae) inferred from chloroplast matK and nuclear
rDNA ITS sequences. Systematic Botany 28: 512-523.
91. Peterson A, John H, Koch E, Peterson J (2004) A molecular phylogeny of the genus Gagea (Liliaceae) in Germany inferred from non-coding
chloroplast and nuclear DNA sequences. Plant Systematics and Evolution 245: 145-162.
92. Utech FH (1992) Biology of Scoliopus (Liliaceae) .1. Phytogeography and Systematics. Annals of the Missouri Botanical Garden 79: 126142.
93. Stevens PF (2008) Angiosperm Phylogeny Website, Version 9, June 2008 (http://wwwmobotorg/MOBOT/research/APweb).
94. Liu Q, Brubaker CL, Green AG, Marshall DR, Sharp PJ, et al. (2001) Evolution of the FAD2-1 fatty acid desaturase 5 ' UTR intron and the
molecular systematics of Gossypium (Malvaceae). American Journal of Botany 88: 92-102.
95. Seelanan T, Brubaker CL, Stewart JM, Craven LA, Wendel JF (1999) Molecular systematics of Australian Gossypium section Grandicalyx
(Malvaceae). Systematic Botany 24: 183-208.
96. Whitlock BA, Bayer C, Baum DA (2001) Phylogenetic relationships and floral evolution of the Byttnerioideae ("Sterculiaceae" or Malvaceae
s.l.) based on sequences of the chloroplast gene, ndhF. Systematic Botany 26: 420-437.
97. Farmer SB, Schilling EE (2002) Phylogenetic analyses of Trilliaceae based on morphological and molecular data. Systematic Botany 27:
674-692.
98. Liden M, Fukuhara T, Rylander J, Oxelman B (1997) Phylogeny and classification of Fumariaceae, with emphasis on Dicentra s.l., based on
the plastid gene rps16 intron. Plant Systematics and Evolution 206: 411-420.
99. Blattner FR, Kadereit JW (1999) Morphological evolution and ecological diversification of the forest-dwelling poppies (Papaveraceae :
Chelidonioideae) as deduced from a molecular phylogeny of the ITS region. Plant Systematics and Evolution 219: 181-197.
100. Rutschmann F, Eriksson T, Abu Salim K, Conti E (2007) Assessing calibration uncertainty in molecular dating: The assignment of fossils to
alternative calibration points. Systematic Biology 56: 591-608.
101. Rutschmann F, Eriksson T, Schonenberger J, Conti E (2004) Did Crypteroniaceae really disperse out of india? Molecular dating evidence
from rbcL, ndhF, and rpl16 intron sequences. International Journal of Plant Sciences 165: S69-S83.
102. Sytsma KJ, Litt A, Zjhra ML, Pires JC, Nepokroeff M, et al. (2004) Clades, clocks, and continents: Historical and biogeographical analysis
of Myrtaceae, Vochysiaceae, and relatives in the Southern Hemisphere. International Journal of Plant Sciences 165: S85-S105.
103. Kathriarachchi H, Samuel R, Hoffmann P, Mlinarec J, Wurdack KJ, et al. (2006) Phylogenetics of tribe Phyllantheae (Phyllanthaceae;
Euphorbiaceae sensu lato) based on nrITS and plastid matK DNA sequence data. American Journal of Botany 93: 637-655.
104. Sutter DM, Forster PI, Endress PK (2006) Female flowers and systematic position of Picrodendraceae (Euphorbiaceae s.l., Malpighiales).
Plant Systematics and Evolution 261: 187-215.
105. Tokuoka T, Tobe H (2006) Phylogenetic analyses of Malpighiales using plastid and nuclear DNA sequences, with particular reference to the
embryology of Euphorbiaceae sens. str. Journal of Plant Research 119: 599-616.
106. Mathews S, Spangler RE, Mason-Gamer RJ, Kellogg EA (2002) Phylogeny of Andropogoneae inferred from phytochrome B, GBSSI, and
ndhF. International Journal of Plant Sciences 163: 441-450.
107. Barker NP, Clark LG, Davis JI, Duvall MR, Guala GF, et al. (2001) Phylogeny and subfamilial classification of the grasses (Poaceae).
Annals of the Missouri Botanical Garden 88: 373-457.
108. Forest F, Chase MW, Persson C, Crane PR, Hawkins JA (2007) The role of biotic and abiotic factors in evolution of ant dispersal in the
milkwort family (Polygalaceae). Evolution 61: 1675-1694.
109. Applequist WL, Wallace RS (2001) Phylogeny of the portulacaceous cohort based on ndhF sequence data. Systematic Botany 26: 406-419.
110. Nyffeler R (2007) The closest relatives of cacti: Insights from phylogenetic analyses of chloroplast and mitochondrial sequences with
special emphasis on relationships in the tribe Anacampseroteae. American Journal of Botany 94: 89-101.
111. Anderberg AA, Trift I, Kallersjo M (2000) Phylogeny of Cyclamen L. (Primulaceae): Evidence from morphology and sequence data from
the internal transcribed spacers of nuclear ribosomal DNA. Plant Systematics and Evolution 220: 147-160.
112. Auld TD, Denham AJ (1999) The role of ants and mammals in dispersal and post-dispersal seed predation of the shrubs Grevillea
(Proteaceae). Plant Ecology 144: 201-213.
113. Barker NP, Weston PH, Rutschmann F, Sauquet H (2007) Molecular dating of the 'Gondwanan' plant family Proteaceae is only partially
congruent with the timing of the break-up of Gondwana. Journal of Biogeography 34: 2012-2027.
114. Hoot SB, Douglas AW (1998) Phylogeny of the Proteaceae based on atpB and atpB-rbcL intergenic spacer region sequences. Australian
Systematic Botany 11: 301-320.
115. Protea_Atlas_Project (2007). pp. Proteaceae - Rarity status, pollination and seed dispersal. Downloaded from
http://protea.worldonline.co.za/default.htm (Accessed Dec 2007).
116. Barker NP, Weston PH, Rourke JP, Reeves G (2002) The relationships of the southern African Proteaceae as elucidated by internal
transcribed spacer (ITS) DNA sequence data. Kew Bulletin 57: 867-883.
117. Johansson JT, Jansen RK (1993) Chloroplast DNA variation and phylogeny of the Ranunculaceae. Plant Systematics and Evolution 187: 2949.
118. Weiss FE (1908) The dispersal of fruits and seeds by ants. New Phytologist 7: 23-28.
119. Paun O, Lehnebach C, Johansson JT, Lockhart P, Horandl E (2005) Phylogenetic relationships and biogeography of Ranunculus and allied
genera (Ranunculaceae) in the Mediterranean region and in the European Alpine System. Taxon 54: 911-930.
120. Manzaneda AJ, Rey PJ, Boulay R (2007) Geographic and temporal variation in the ant-seed dispersal assemblage of the perennial herb
Helleborus foetidus L. (Ranunculaceae). Biological Journal of the Linnean Society 92: 135-150.
121. Moline PM, Linder HP (2005) Molecular phylogeny and generic delimitation in the Elegia group (Restionaceae, South Africa) based on a
complete taxon sampling and four chloroplast DNA regions. Systematic Botany 30: 759-772.
122. Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD (2004) Historical biogeography of two cosmopolitan families of flowering
plants: Annonaceae and Rhamnaceae. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 359:
1495-1508.
123. Anderson CL, Rova JHE, Andersson L (2001) Molecular phylogeny of the tribe Anthospermeae (Rubiaceae): Systematic and biogeographic
implications. Australian Systematic Botany 14: 231-244.
124. Andersson L, Rova JHE (1999) The rps16 intron and the phylogeny of the Rubioideae (Rubiaceae). Plant Systematics and Evolution 214:
161-186.
125. Auld TD (2001) The ecology of the Rutaceae in the Sydney region of south-eastern Australia: Poorly known ecology of a neglected family.
Cunninghamia 7: 213-239.
126. Mole BJ, Udovicic F, Ladiges PY, Duretto MF (2004) Molecular phylogeny of Phebalium (Rutaceae: Boronieae) and related genera based
on the nrDNA regions ITS 1+2. Plant Systematics and Evolution 249: 197-212.
127. Pilger R (1935) Santalaceae. In: Engler A, Prantl R, Harms H, editors. Die Naturlichen Pflanzen Familien. 2nd ed. ed. pp. 52-91.
128. Der JP, Nickrent DL (2008) A molecular phylogeny of Santalaceae (Santalales). Systematic Botany 33: 107-116.
129. Reynolds ST (1981) Austrobaileya 14: 388-419.
130. Harrington MG, Edwards KJ, Johnson SA, Chase MW, Gadek PA (2005) Phylogenetic inference in Sapindaceae sensu lato using plastid
matK and rbcL DNA sequences. Systematic Botany 30: 366-382.
131. Hughes L, Westoby M (1990) Removal rates of seeds adapted for dispersal by ants. Ecology 71: 138-148.
132. Jurado E, Westoby M, Nelson D (1991) Diaspore weight, dispersal, growth form and perenniality of central Australian plants. Journal of
Ecology 79: 811-830.
133. Fischer E (2004) Scrophulariaceae. In: Kubitzki K, Kadereit JW, editors. The Families and Genera of Vascular Plants Vol 7, Flowering
Plants Dicotyledons: Lamiales (except Acanthaceae including Avicenniaceae). Berlin: Springer. pp. 333-432.
134. Bennett JR, Mathews S (2006) Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. American Journal of
Botany 93: 1039-1051.
135. Wolfe AD, Randle CP, Liu L, Steiner KE (2005) Phylogeny and biogeography of Orobanchaceae. Folia Geobotanica 40: 115-134.
136. Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, et al. (2008) A molecular phylogeny of the Solanaceae. Taxon 57:
1159-1181.
137. Cuautle M, Rico-Gray V, Diaz-Castelazo C (2005) Effects of ant behaviour and presence of extrafloral nectaries on seed dispersal of the
Neotropical myrmecochore Turnera ulmifolia L. (Turneraceae). Biological Journal of the Linnean Society 86: 67-77.
138. Schappert PJ, Shore JS (2000) Cyanogenesis in Turnera ulmifolia L. (Turneraceae): II. Developmental expression, heritability and cost of
cyanogenesis. Evolutionary Ecology Research 2: 337-352.
139. Truyens S, Arbo MM, Shore JS (2005) Phylogenetic relationships, chromosome and breeding system evolution in Turnera (Turneraceae):
Inferences from its sequence data. American Journal of Botany 92: 1749-1758.
140. Sytsma KJ, Morawetz J, Pires JC, Nepokroeff M, Conti E, et al. (2002) Urticalean rosids: Circumscription, rosid ancestry, and
phylogenetics based on rbcL, trnL-F, and ndhF sequences. American Journal of Botany 89: 1531-1546.
141. Hidalgo O, Garnatje T, Susanna A, Mathez J (2004) Phylogeny of Valerianaceae based on matK and ITS markers, with reference to matK
individual polymorphism. Annals of Botany 93: 283-293.
142. Tokuoka T (2008) Molecular phylogenetic analysis of Violaceae (Malpighiales) based on plastid and nuclear DNA sequences. Journal of
Plant Research 121: 253-260.
143. Beier BA, Chase MW, Thulin M (2003) Phylogenetic relationships and taxonomy of subfamily Zygophylloideae (Zygophyllaceae) based on
molecular and morphological data. Plant Systematics and Evolution 240: 11-39.
144. Sheahan MC, Chase MW (2000) Phylogenetic relationships within Zygophyllaceae based on DNA sequences of three plastid regions, with
special emphasis on Zygophylloideae. Systematic Botany 25: 371-384.