TheCellLineOntology
advertisement

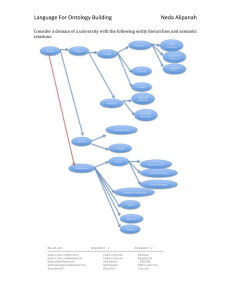
1)National Center for Integrative Biomedical Informatics and the Center for Computational Medicine and Biology 2)Department of Psychiatry, University of Michigan, Ann Arbor, Michigan 48109 3)Department of Human Genetics, University of Michigan, Ann Arbor, Michigan 48109 The Cell Line Ontology Sirarat Sarntivijai, Alexander S. Ade, Brian D. Athey ,and David J. States Abstract Cell lines are used extensively throughout biomedical research, but the nomenclature describing cell lines has not been standardized, and many ambiguous or non-unique names appear in the published literature. The Cell Line Ontology is a well-structured collection of names for cell lines cultured in vitro with descriptive data. This ontology contains a broad collection of cell line names compiled from ATCC, Hyper CLDB, MeSH and primary literature scanning. In addition to names, the ontology specifies relationships between cell lines including derivation and homology. The Cell Line Ontology facilitates data exploration and comparison of different cell lines in support of clinical and experimental research. We analyze the use of cell line names in biomedical text. Issues that are encountered when we attempt to interpret such tokens in natural language processing include ambiguous names, polymorphisms in the use of names, and the fact that some cell line names are also common English words. Linguistic patterns associated with the occurrence of cell line names are analyzed. Applying these patterns to find additional cell line names in the literature, identifies only a small number of additional names. Annotation of microarray gene expression studies is used as a test case. The objectives of this work are (a) to assist users in extracting useful information from biomedical text and (b) to highlight the importance of standardizing cell line names in biomedical research. Background and Problem Cells cultured in vitro are powerful and convenient model systems that are widely used in biomedical research. As they are developed at lab benches, cell line names are assigned by their laboratory of origin, or sometimes, a catalogue number is added when they are submitted to a repository. Cell line names are inherited, reproduced, or preferentially formatted with variations in the published literature. Non-unique or ambiguous names also appear in the literature. The inconsistency and ambiguity in misleading or non-informative cell line names add complications to data-mining and knowledge-seeking activities. Here, we propose the Cell Line Ontology that includes structured knowledge about cell lines and their derivatives and homologues. Methods Large collections of cell line information were obtained from American Type Culture collection (ATCC) catalogue (available online at http://www.atcc.org/common/documents/pdf/CellCatalog/CellIndex.pdf as of November 2006) and Hyper Cell line Data Base (CLDB) version 4.200201 (http://www.biotech.ist.unige.it/interlab/cldb.html). Cell line names in the NLM Medical Subject Headings (MeSH) are linked to their corresponding cell line. Name duplication across the data sources is eliminated while the provenance of those duplicates is kept in the cross-reference links for each cell line. After data cleaning, we found 8,740 cell line instances. Each cell line record holds the information attributes of CellLineID, Organism, Tissue, Pathology, Growth Mode, Repository Source, ATCC number, CLDB html, and MeSHID. ATCC number is the ATCC catalogue number, and CLDB html identifier is the orginal html file names as taken from the base URL (http://www.biotech.ist.unige.it/cldb/ *.html). The ontology was constructed using Stanford Protégé ontology editor. A Java script was written to automate data loading to this ontology in a W3C Web Ontology Language Format (OWL-DL). • Data (Preprocessing) – ATCC • .PDF catalogue • 3,489 distinct catalogue numbers – Hyper CLDB • .html web pages • 6,856 entries with duplicates -> 5,889 case-sensitive unique name entries. – Total ATCC + CLDB • 8,861 non-duplicate non-overlap entries -> 8,740 entries after data cleaning • Ontology Creation – Ontology structure constructed in Protégé ontology editor – Java script for automating instance creation in a W3C Web Ontology Language Format (OWL-DL) Results Most of the cell line entities in this Cell Line Ontology are stand-alone instances with the link to their source of origin. However, a number of cell lines are derivatives from some common cell lines. These entities contain the isDerivedFrom relationship to their parents. Cell lines that are parents of some derivatives also contain the hasDerivatives relationship to their children cell lines. For cell lines that derive from a common parent cell line, they have transitive hasHomolog relationship to their siblings (other clones from same parent). With the creation of this ontology, we have also witnessed the use of cell line names that are non-unique, extremely common, and ambiguous throughout the biomedical text. We have performed an experiment using Natural Language Processing (NLP) approach to evaluate these cell line names. Noun phrases ending in ‘cells’ or ‘cell line’ are likely to refer to cell lines and can help in tagging cell line names that are also common English words, but requiring the use of these constructs impairs recall in named entity recognition. Data Structure UML Diagram describing the entities and relationships of the Cell Line Ontology Top ten Hyper CLDB cell line names that appear in non-biomedical corpus ranked by number of occurrences Information Access through the Cell Line Ontology • Genotypic information – Next release: breast cancer genotypic information from Lawrence Berkeley National Laboratory • Quick cell line information in PubMed without the hassles – NLP implementation killing false positives of the short or common-english-word cell line names. Cell Line Ontology Examples of Cell Line Synonyms Hyper CLDB cell line names that appear in NCBI GEO Microarray sample description, ranked by number of occurrences. Top ten Hyper CLDB cell line names that appear in non-biomedical corpus ranked by number of occurrences Cell Line Name Count M4 48 35 44 Aa 11 EC 11 P1 11 380 9 BT 8 L 7 CAR 4 OK 4 Conclusion The non-standardized nomenclature of cell line names has brought several issues in NLP to attention. Ambiguity in the use of these names needs to be resolved wherever possible. The issues includes the use of special characters in Unicode encodings, inconsistent format in punctuation marks, the use of common names (e.g. HORSE, MOUSE, OK, L and 35 for cell lines). The Cell Line Ontology is the first step to aid cell line information mining while these annotation issues are not yet resolved at the present. References • Bard, J. Rhee, S.Y. Ashburner, M. (2005) An ontology for cell types Genome Biol., 6(2): R21 • Frank, E. Hall, M. Trigg, L. Holmes, G. Witten, I.H. (2004) Data mining in bioinformatics using Weka Bioinformatics, 20(15), 2479-2481 • Lee, K.J. Hwang, Y.S. Kim, S. Rim, S.C. (2004) Biomedical named entity recognition using twophase model based on SVMs J. Biomed Inform., 37(6), 436-447 • Liu, H. Hu, Z.Z. Torii, M. Wu, C. Friedman, C. (2006) Quantitative assessment of dictionarybased protein named entity tagging J. Am. Med. Inform. Assoc., 13(5), 497-507 • NLM (2007) Medical Subject Headings http://www.nlm.nih.gov/mesh/meshhome.html • Neve, R.M., et al. (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes, Cancer Cell. 2006 Dec; 10(6): 515-27. • Noy, N.F. Sintek, M. Decker, S. Crubezy M. Gerguson, R.W. Musen, M.A. (2000) Creating Semantic Web Contents with Protégé-2000 Intelligent Systems, IEEE Vol. 6, 2, 60-71. • Noy, N.F. Crubezy M. Gerguson R.W. Knublauch, H. Tu, S.W. Vendetti, J. Musen, M.A. (2003) Protégé-2000: an open source ontology-development and knowledge-acquisition environment AMIA Annual Symposium Proceedings 2003, 2003: 953 • Rinaldi, F. Schneider, G. Kaljurand, K. Hess, M. Romacker, M. (2006) An environment for relation mining over richly annotated corpora: the case of GENIA BMC Bioinformatics, 2006: 7(Suppl 3):S3 • Shulz, S. Beisswanger, E. Wermter, J. Hahn, U. (2006) Towards an upper level ontology for molecular biology AMIA Annual Symposium Proceedings 2006, 694-698 • University of Illinois at Urbana Champaign (2004) Sentence segmentation tool http://l2r.cs.uiuc.edu/~cogcomp/atool.php?tkey=SS Acknowledgements The authors thank Mark Musen and Natasha Noy at Stanford BMI for Protégé support, and Chachrist Srisuwanrat for VBA technical support. This work has been done with the support from NIH under grant U54 DA021519 for the National Center for Integrative Biomedical Informatics and R01 LM008106. The web ontology file for this cell line collection can be downloaded at http://www.stateslab.org/data/celllineOntology/cellline.zip