We present and compare the results of a partial QTL detection for
advertisement

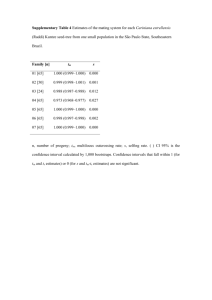
Abstract We present and compare the results of a partial QTL detection for fertility in a table grape progeny, MTP3346. Novel parental and consensus maps were built, spanning ten genomic regions. The study of this progeny essentially confirmed the most stable QTL found on LG 5 in the two other progenies described in the manuscript, MTP3140 and MTP3234. Material and methods The MTP3346 progeny consisted of 519 offspring from a cross between one seeded cultivar (Muscat of Alexandria) and one seedless individual (MTP3140-517, selected from the MTP3140 progeny). It was initially intended to study seedlessness inheritance. MTP3346 plants were grown in containers in a plastic greenhouse, with each cane left unpruned and wound up like a hunting horn, at the CTIFL experimental station of Balandran, Gard, France in 2001-2003. In 2004, a random subset of 142 fertile plants plus a random subset of 20 non-fertile plants were installed outside in containers at the Chapitre experimental station, to check the results obtained in 2003. MTP3346 was left unpruned, because low fertility was expected on basal shoots in the first years after plantation due to the seedless parentage of this progeny. Fertility was measured in 2003 and 2005. Only inflorescences on the two terminal shoots were counted because not all shoots could be left on the cane due to the high plant density within the greenhouse. Since cane diameter and terminal shoot rank may be related to shoot fertility (Huglin 1958) and these traits were highly variable within this progeny, we recorded them too. In addition to normality of the distributions, phenotypic correlations, and year effects, the effects of cane diameter and terminal shoot rank were also tested using SAS 8.1 software. For MTP3346, maps were constructed for ten genomic regions only, with 29 SSRs (2 to 4 per region), because this mapping work was initially intended for checking QTLs detected for seedlessness and phenology in MTP3140. All 519 individuals were genotyped to build the maps. SSR genotyping was performed as described in Doligez et al. (2006b). For MTP3346, complete and framework genetic maps were constructed as for MTP3140, with a maximum recombination rate threshold of 0.4 for parental maps. QTL detection was performed as for MTP3140 and MTP3234, including CIM detection even though the choice of cofactors might be affected due to the partial coverage of genetic maps. Results The raw distributions significantly deviated from normality (P < 0.0001). However, the distribution was not skewed apart from a large zero class in 2003 (Fig. S1), and applying the square root or the neperian logarithm functions to the raw data did not yield normality. Therefore, we did not apply any transformation to the raw data. Phenotypic correlation between years and year effect were significant (Table S1). The linear regression of raw fertility on cane diameter and/or terminal shoot rank for MTP3234 and MTP3346 yielded highly significant results, but the R2 values were low (maximum 0.14, data not shown). The partial maps which were built in this study are shown in Fig. S2. The effect of cane diameter and/or terminal shoot rank on raw fertility was significant but weak (see above). Moreover, QTLs were found in the same genomic regions and explained equivalent percentages of variance, when using the residuals of the regression models instead of the unadjusted fertility data in QTL detection (data not shown). Therefore, we used unadjusted fertility data in the following QTL analyses. The main QTL found in progenies MTP3140 and MTP3234 on one end of LG 5, near SSR marker VVMD27, was also present in the two parents of progeny MTP3346, but in one year only (Table S2 and Fig. S3). The second QTL found on LG 5 in progeny MTP3234, close to the main one, around marker VVIT68, was also found in MTP3346, but in one parent and one year only. In both parents, the residual distribution of the CIM model at QTLs on LG 5 departed significantly from normality (Table S2). This might result from missing cofactors due to the partial genetic maps. However, these QTLs were confirmed through single marker analysis of variance, which was significant at = 0.01% or 0.1% for SSRs VVMD27, VRZAG79 and VVIT68 in both parents. For the female parent, two LOD peaks were obtained on LG 5 with a 10 cM cofactor exclusion window, but only the peak near VVMD27 remained when reducing this window to 1 cM (data not shown). Discussion This partial QTL detection also revealed the same main QTL for fertility found in the two progenies MTP3140 and MTP3234 on LG 5, despite the intrinsic limitations of a search for QTLs with partial maps and despite differences in vine training (pruning), environmental conditions (years, field vs plastic greenhouse) and trait measure (shoot rank on cane). Higher LOD scores were found in 2003 in MTP3346 than in the two other progenies probably due to the much larger progeny size (519). The second QTL of LG 5 was also found in MTP3346. Unfortunately, since only partial maps were built for MTP3346, the comparison with this progeny could not be extended to all genomic regions. In particular, the partial maps of MTP3346 did not cover the region of LG 14 where stable QTLs were found in MTP3234. The range of fertility values in MTP3346 was very similar to MTP3140 and MTP3234, although MTP3346 harboured alleles from the seedless cultivar Sultanina (Thompson seedless). The low fertility expected for this reason was balanced by long pruning and measuring fertility on terminal rather than basal shoots, which allowed to reach fertility values similar to those in the progeny MTP3234 without seedless ancestry and to identify a common QTL on LG 5. We found higher phenotypic correlations for fertility between years in progeny MTP3346 than Fanizza et al. (2005) for the number of clusters per vine. In MTP3346 the rather high phenotypic correlation is not consistent with the lack of stability over years of the QTL on LG 5. Since this progeny was observed in very different environments in 2003 (greenhouse) and 2005 (outside), phenotypic correlation in this progeny is probably not mainly environmental. Rather, the decrease in QTL detection power due to the reduced progeny size is probably responsible for the lack of QTL stability. The MTP3346 progeny size was very similar in 2005 to the progeny sizes of MTP3140 and MTP3234. But the contrasts between alleles at the main QTL on LG 5 may be lower in MTP3346 than in MTP3140 and MTP3234, which could explain that this QTL was detected with the very large progeny size in 2003 but not with the smaller progeny size in 2005. For MTP3346, the fact that QTLs were found only in 2003, concomitant with the occurence of a large zero class in this young progeny, is suggestive of some relationship with the length of juvenile period, which is cultivar-dependent. However, QTLs were still found on LG 5 in 2003 after deleting the zero fertility class (data not shown). Therefore, the lack of QTL detection in 2005 seems to be a consequence of the progeny size reduction between 2003 and 2005, leading to reduced QTL detection power, rather than a consequence of the drastic reduction of the zero class between 2003 and 2005. Table S1. Year effect on fertility and phenotypic correlations between years, in three table grape progenies. Progeny Years compared Year effect Spearman correlation MTP3346 2003-2005 P = 0.0031 0.45 (P < 0.0001) Table S2. QTLs for fertility detected in three table grape progenies, with several methods. Progeny Mapa Year LGb MTP3346 KWc F F 2003 5 2003 5 VVMD27, VrZAG79 VVIT68 M 2003 5 VVMD27, VrZAG79, VVIT68 IMd LOD LOD % additive confidence threshold peak variance effect intervalf 2.1 7.40 6.7 0.41 22.2 - 29.4 2.1 2.1 8.19 7.4 0.42 16.5 - 23.8 LOD threshold 2.6 2.6 LOD peak 9.77 3.37 5.3 7.47 CIMe % additive confidence variance effect interval Sg 9.5 -0.51 23.4 - 29.4 11.58 ** 4.3 0.36 11.3 - 14.2 12.49 ** 6.5 0.40 19.8 - 23.8 14.57 ** C 2003 4 3.2 3.2 h 3.33 3.4 11.9 - 15.9 20.1 - 27.4 C 2003 5 VVMD27, VrZAG79, VVIT68 3.2 19.47 16.9 3.2 h 19.73 14.6 20.1 - 27.4 h C 2003 12 3.2 4.42 6.2 0.0 - 14.7 3.2 4.29 4.2 0 - 14.7 a F: female, M: male, C: consensus b LG: linkage group c Loci for which KW (Kruskal-Wallis) test was significant at = 0.001 d IM: interval mapping e CIM: composite interval mapping f Position of one-LOD support confidence intervals on LGs, in cM g S: statistics for the test of normality of residual distribution computed in QTL Cartographer (**: P < 0.01, ns: P 0.05) for CIM h IM LOD thresholds (in the absence of implemented permutation test for consensus maps) Note: KW, IM, and consensus CIM results were obtained with MapQTL. Parental CIM results were obtained with WinQTLCart. For progeny MTP3346, QTLs are for raw data. - Fig. S1. Distribution of raw fertility measures in table grape progeny MTP3346. x axis: classes of fertility values, y axis: number of individuals of the progeny in each fertility class. n: number of individuals scored. MTP3346 2003 (n = 491) 2005 (n = 151) 120 100 80 60 40 20 0 0 0.5 1 1.5 2 2.5 3 Fig. S2. Parental and framework genetic maps for 10 genomic regions of the table grape progeny MTP3346. In the very few cases for which framework maps differed from complete maps, only framework maps are shown. Distances are in cM Kosambi. F: female, C: consensus, M: male, fw: framework. Stars indicate significantly distorted loci (* P < 0.05, ** P < 0.01, *** P < 0.001). LG 4 F LG 5 Cfw F M Cfw VRZAG83 0.0 11.9 15.9 VMC2B5F LG 7 0.0 VVMD32 VMC2B5M F M VMC1E11AF C 0.0 LG 8 M F VVIV04 VVIT68 11.3 VRZAG79 VVMD27 20.1 27.4 14.8 VMC1A12 35.6 VMC8D11 C M 0.0 VMC6G8 *** *** 9.8 16.5 VMC2F12 *** VMC1F10 *** *** *** LG 16 LG 12 F LG 11 C * Cfw Mfw C 0.0 F M VMC6C10 19.7 VMC2H4 34.3 VMC8G6 * 16.5 VMC1E12 * 36.4 VMCNG1E1 ** Cfw 0.0 20.6 LG 17 0.0 6.0 32.0 LG 18 M C VVIB09 F C 0.0 VVIN16A 25.9 VMC7F2 VMC9G4 VVIQ22.1 VMC1E11B * * 48.9 VVMD37C F VVIN52 * * VVMD25 17.5 M F VMCNG2D11 0.0 VVIB19 0.0 LG 14 VVMD37A Mfw Fig. S3. One-LOD support confidence intervals of the main QTLs for fertility detected with CIM on framework maps in the table grape progeny MTP3346. F: female, C: consensus, M: male. QTLs on parental maps were detected with WinQTLCart and QTLs on consensus maps with MapQTL. QTLs on consensus maps are represented only when their LOD peak was larger than the IM LOD threshold + 0.5. QTLs are for raw data. MTP3346 – LG 5 F 0.0 C 0.0 VMC1E11AF 11.3 VVIT68 22.2 VrZAG79 29.4 VVMD27 2003 LOD 3.37 2003 LOD 9.77 M VMC1E11aF 11.3 VVIT68 20.1 VrZAG79 27.4 VVMD27 0.0 VMC1E11aM 9.7 VVIT68 2003 LOD 19.73 16.5 VrZAG79 23.8 VVMD27 2003 LOD 7.47