In situ hybridization protocol
advertisement

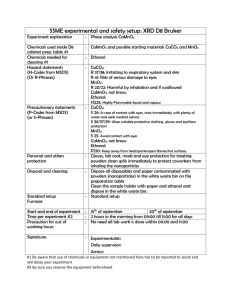
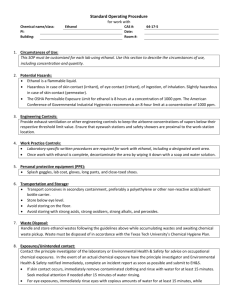
CH in situ protocol January 2011 1 In situ hybridization protocol (Modifications for Liu Lab based on Bob Frank’s and Chloe/Vivian Irish’s versions) Part I: Embedding and sectioning 1. Follow embedding protocol by Drews, Jack, and Weigel modified by Meyorowitz lab 2. Prepare slides with 8m sections of tissue in correct orientation by placing pieces of sectioned ribbon onto slides covered in RNAse-Free water on 42o C slide warmer. Label slides with pencil. Leave slides on slide warmer until dry (Usually they are left overnight, but if embedding isn’t perfect and slides are left too long the bubbles may become apparent and tear apart the tissue). Part II: Non-Radioactive Probe Synthesis (from plasmid) For this, you first need to bulk up your plasmid containing the probes. A midi prep is best, which can give you ~1ug/ul Be sure to use RNAse-Free filter tips, Rnase-Free solutions, clean pipets with RNAseaway, and be extra clean and aware of possible RNAse contamination 1. Using appropriate enzyme to linearize plasmid, digest overnight 5-15g of plasmid in a 50 l rxn. (Note: Enzyme must leave a 5’ overhang) 2. Run a gel with ~1ul sample to check band and visualize approximate concentration 3. Add 450 l 1X TE to digest to bring total volume up to ~500 l 4. Perform a Phenol:Chloroform purification of Linear DNA a. Add 500 l Phenol:Chloroform (or an equal amount to volume of sample) b. Vortex to mix well c. Spin (~13,000 rpm) 8 minutes. Room temp is okay. d. Carefully transfer top layer into an RNAse-Free tube e. Add 50l (or 1/10 volume) of sterile 3 M Sodium Acetate, pH ~5.2 (The addition of this neutralizes the solution and the salt associates with the negatively charged DNA and allows it to precipitate) f. Add 2-2.5 volumes (1000l) of 100% EtoH g. Leave at -20oC for 2 hours (more time is better) h. Spin 20 minutes 4oC (~13,000rmp) i. Decant ethanol and wash pellet with cold 70% ethanol j. Air dry pellet k. Resuspend in 20 l Rnase-Free water, quantify DNA with spec or nanodrop. Store -20oC if continuing the next day 5. Perform non-radioactive labeling rxn following a modified version of ROHE DIG RNA labeling protocol ( cat 11-277-073-910). This can be done the day after purification. a. In a RNAse-Free tube, combine: CH in situ protocol January 2011 2 l (1 g) linear DNA 2 l DIG labeling mix (Roche Cat No 11 277 073 910) 2 l 10x T7 Buffer l RNase-Free (Milli-Q) dH20 1 l RNase-Out (RNAse inhibitor – can use the one in RT kit) 2 l T7 RNA polymerase (Roche 10 881 767 001) 20 l final volume b. Incubate at 37oC for ~2hrs (company protocol is 2hrs, Chloe does 4) c. Add 1l more of T7 RNAse polymerase and incubate another 2 hours at 37 oC (can shorten time; this is not in company protocol) d. Add 2l DNAse I and incubate 37oC for 15-30 minutes e. Remove from 37o C and add 2 l 0.2M sterile EDTA to stop rxn f. Run 1-2 l of RNA sample on a gel with 100 bp DNA ladder. Run fast (~125V) so that gel will be warm and RNA is less likely to form secondary structures. Note: RNA size compared to DNA ladder will not be exact and it is likely that several bands may be visible. You may want to heat the RNA for 5 min at 80 deg before running on gel. g. Estimate concentration and total amount of RNA probe by comparing to ladder concentrations 6. Perform an ethanol precipitation of RNA transcript (to clean up RNA) If using “B” tube from -80, start at step d a. Add 5 l of sterile 4M LiCl and 150 l of 100% Ethanol to RNA transcript b. Split volume between two RNAse-Free tubes (~90l each, I call them ‘A’ and ‘B’). Note how much RNA is in each tube, according to estimate from gel. Leave the ‘B’ tube in the – 80 o C freezer for future use. (This step is done to save extra probe in a stable state. It does not need to be done. All of the following steps can be done with full volume of probe) c. Leave ‘A’ tube (or original tube, if not splitting) in – 20o C freezer overnight for RNA precipitation d. The next day, spin tube at 4 o C, 20 minutes (~13,000 rpm) e. Decant ethanol and wash pellet with 1ml cold 70% Ethanol made with DEPC water f. Spin 5 minutes. Air dry (or speedvac with no heat) pellet g. Dissolve in 30l RNAse-Free water i. Set aside 2.5 -3 l for quantifying on the nanodrop and running on a gel before hybridization. Heat RNA for gel at 80deg for 5 min. h. Store in -80o C freezer Part III: Wax removal and Hybridization Prepare the following ahead of time: CH in situ protocol January 2011 3 Bake: slide rack and handle, 17 glass boxes (we only have 18 left, so be gentle with them), one 500 ml flask and one regular/medium sized stir bar for making paraformaldehyde solution, glass graduated cylinders (two 500ml, one or two 250 ml, and one 100m if you have onel), a few razor blades and flat spatulas. And a funnel or two. It is also good idea to bake an extra 500ml flask, a medium sized stirbar, and a 1L bottle in case mistakes are made. Autoclave: 7 liters of autoclaved dH20 in baked bottles (This is more than enough for the whole experiment Make 10X in situ salt solution and store 1 ml aliquots in – 20oC freezer This is needed to make the hybridization solution 6 ml 5M NaCl 1 ml Tris pH 6.5 (sterile) 2 ml 0.5M NaPO4 buffer pH 6.8 (containing both mono-and di-basic forms) 1 ml 0.5 M EDTA 10 ml Make Hybridization solution (without tRNA) and store in 1.5 - 2ml aliquots in -20o C. 80 l of Hyb solution are needed per slide. A 2ml aliquot is good for up to a total of ~23 slides, and a 1.5 ml aliquot is good for 18 slides per probe For 8 ml (minus volume for tRNA) of Hybridization solution, add Make dextran with water. Freeze aliquots in -20 1 ml 10 X in situ salts - needs to be made, not bought, store aliquots in -20o 4 ml formamide (stored in -20) 2 ml 50 % Dextran Sulfate (Very viscous. When making, heat at 60o ) 200 l 50X Denhardts solution 700 l RNAse-Free (Milli-Q or DEPC) water 7 mls 900 l On day of experiment, add fresh tRNA to tubes (for 2 ml aliquots, add 25 l of 100mg/ml tRNA solution, for 1.5 ml aliquots add 19 l – this gives 1 l for every possible 80l rxn) The tRNA is Roche cat 109517, made up with mqDH2O and frozen in aliquots of 50l. Make 50% Formamide 1ml aliquots and store in -20o C. Make sure the following reagents are available: Glycine (1g), paraformaldehyde (14g), blocking reagent (1g), BSA (5g) 100% EtOH – 4 Liters 10X PBS (DEPC treated) – need at least 200 ml 20X SSC (DEPC treated) – at least 45 ml 1M Tris-HCl pH 8, sterile – at least 35 ml 0.5M EDTA, sterile – at least 35 ml CH in situ protocol January 2011 4 1M Tris-HCL pH 7.5, sterile – at least 60 ml 1M Tris-HCl pH 9.5, sterile - at least 20 ml 5M NaCl, sterile – at least 22 ml 2M MgCl2 , sterile – at least 5 ml Triton X-100 – at least 1.5ml Western Blue Proteinase K (Sigma-2308), 10 mg/ml, at least 125 l Anti-DIG antibody (8 l) Reserve incubator for first two days of hybridization Set up 37oC water bath. 1st Day of Experiment: Preparation: The first day takes ~ 4+ hrs (including time for making solutions), depending on how many samples and probes are involved 1. Turn on and set incubator to ~52oC (depending on decided hybridization temperature. Higher temp will be more stringent) 2. Set up/turn on 37oC water bath 3. Pre-warm Hyb solution aliquot in ~52oC incubator 4. Prepare 2 L of 1x PBS (make 2 1L bottles, 100ml of 10X DEPC treated PBS each bottle + water to fill to 1 L) * Make 1 Liter at a time, using one of the bottles containing autoclaved water * 5. Prepare 350ml 4% paraformaldehyde in 1X PBS (14g for 350 ml) a. This is a very toxic chemical. Weigh in hood, or wear mask. Use special waste container when done with this at the end of the day. b. To dissolve, add 30 to 60ull of 5M NaOH or one chip of NaOH and stir with heat on high (this can take over an hour, and then it needs time to cool!!!) 6. Prepare individual glass boxes with 350ml of the following solutions: a. 2 x 100 % xylene. (Add small stir bar and leave in hood) b. 2x 100% ethanol. (Leave in hood) c. 1 x 95 % ethanol (332.5 ml 100% EtOH + 17.5 ml dH2O) d. 1 x 90% ethanol (315 ml 100% EtOH + 35 ml dH2O) e. 1 x 80% ethanol (280 ml 100% EtOH + 70 ml dH2O) f. 1 x 60% ethanol (210 ml 100% EtOH + 140 ml dH2O) g. 1 x 30% ethanol (105 ml 100% EtOH + 245 ml dH2O) h. 2 x 100% dH2O i. 1 x 2X SSC (35 ml of 20X SSC + 315 ml CH in situ protocol January 2011 5 j. 1 x 100 mM Tris pH8, 50 mM EDTA (35 ml of 1 M Tris pH8 + 35 ml 0.5M EDTA + 280 ml water). Place this box in the 37o water bath to equilibrate. Later, you willl add 125 l of 10mg/ml proteinase K immediately before using!!!! k. 1 x 2mg/ml glycine in 1X PBS ( add 0.7g glycine to 350ml 1X PBS) l. 3 X 1X PBS (label 1,2, and 3) m. 1 x 4% paraformaldehye in 1X PBS (made above). Leave in Hood! 7. Set heat block to 80o (Can do during proteinase K incubation) Day 1 Protocol: With slides in rack, do the following washes/incubations 1. Ethanol dehydration (Room temperature) a. b. c. d. e. f. g. h. 2 washes in Xylene 10 minutes each 2 washes in 100% ethanol (1st wash 2 minutes, second wash 1 minute) 95% ethanol (2min) 90 % ethanol (2 min) 80 % ethanol (2 min) 60% ethanol (2 min) 30% ethanol (2 min) 2 washes in dH20 (2 min, 1 min) 2. One wash with 2X SSC (15-20 min) at room temperature a. This box will be used again on day 2 with dilute 0.2x SSC 2. Proteinase K (just before incubation, add prot k for a final conc of 0.75-1 ug/ml (35 ul of 10mg/ml prot K for 1ug/ul) to the box in the 37oC waterbath containing 350ml 100mM Tris 8, 50 mM EDTA). a. Make sure liquid in box is 37 deg! The water bath may have to be ~40 for the box to be 37! b. Incubate in 37oC water bath for 30 minutes, gently dunking slide rack up and down every 5-10min (Gentle shaking in 37o dry incubator can be done instead) c. The time and amount of proteinase K can be decreased if too much tissue degradation occurs d. During this time, set a heat block to 80o C 3. 2mg/ml glycine in1X PBS (2 min) (can go a little longer, Bob does 5 min) 4. 2 washes of 1X PBS (2 min each) a. uses boxes labeled 1 and 2 5. 4 % paraformaldehyde, in hood (10 minutes) - CH in situ protocol January 2011 6 a. While waiting, set up Tupperware box for hybridization: clean with RNAse away, rinse with autoclaved water, cover bottom with paper towels, wet, pour off extra water, lay down serological pipets that will be used as a rack for slides, and wrap lid with plastic wrap to ensure a tight seal. 6. 1X PBS (2 min) -**Reuse PBS box 2** 7. 1X PBS (2 min) **Use box 3, and then save it for day 2** 8. Ethanol Dehydrations, 30 seconds each a. b. c. d. e. f. g. 30% 60% 80% 90% 95% 100% (reuse EtOH box 2) 100% Fresh solution!!! (Empty one of the dH2O boxes and fill with EtOH) 9. Temporarily store slides in box with a little bit of ethanol (not submerged) while preparing hybridization master mixes 10. In a RNAse-Free Tube, combine appropriate amounts of probe and 50% formamide for the number of slides you have plus one: a. Final volume per slide should be 20l (ie 1 l probe + 19 l 50 % formamide) b. If you have 16 slides, make mix for 17, so you extra to account for loss from pipetting 11. Heat probe/formamide mix for 2 minutes at 80o. Immediately cool in ice water when done. 12. Add tRNA to pre-warmed hyb solution (that was warming in the ~52o incubator) a. 19 l tRNA for 1.5 ml hyb solution aliquot b. 25 l tRNA for 2 ml hyb solution aliquot 13. To the RNA probe/Formamide mix, Add 80 l of Hyb solution (containing tRNA) for every slide. (i.e. If you have 15 slides, and made mix for 16 slides, add 80l x 16 = 1280 l hyb solution to the mix) 14. Remove slides from rack and set on paper towels to dry 15. Add 100l of hyb mix (containing probe, formamide, and hyb solution with tRNA) to each slide. Add by dropping ~4 drops across the slide. Change gloves btwn probes 16. Carefully lower coverslip onto slide, so that hyb mix spreads with out air bubbles CH in situ protocol January 2011 7 17. Place slides onto (pipet) rack in Tupperware box. Seal, and leave overnight in ~52o incubator (higher temp is more stringent/ (Bob does 36 hrs, 56 deg!) 18. Prepare 0.2X SSC for next day, and leave in 52o incubator overnight a. Need 800ml (2 x 350ml boxes, 50ml for dipping slides) b. 8 ml 20X SSC + 792 ml water c. Store in one of the emptied autoclaved water bottles 19. Save PBS box # 3 with some liquid in it for Day 2 Day 2 Protocol: (takes about 7.5 – 8 hrs) 1. Remove cover slips from slides by a combination of hanging them vertically, carefully using a (baked) blade, and dipping them into a 50cc tube of 0.2X SSC (which was warmed overnight), and return slides to metal slide rack. Change SSC btwn probes. 2. Wash slides two times, for one hour each, in 0.2X SSC shaking in ~52o. a. Use the SSC box from day 1, after rinsing, for both washes. After first wash, pour out solution from the first wash and replace with fresh 0.2X SSC for second wash. b. During the first wash, the bottle containing the remaining 0.2X SSC should be returned to the incubator. c. Do not wash for longer than the two hours!!!! d. During these washes, make up the following solutions Note: This blocking solution may not be best!!! Might want to revert to original method, (or variation of it with out the acid) from manufactures protocol!!!! Blocking Solution: (Need 75- 100 ml, but easiest to make up 2 x 50 ml tubes) 1.5 ml 5M NaCl 5 ml 1M Tris pH 7.5 0.5 g Blocking reagent (Roche 11 096 176 001) autoclaved dH2O to 50ml (remember to make two tubes!) Place Blocking solution in 60o incubator to dissolve. This will take ~1hr!! Make the following in now empty autoclaved water bottles BSA/Tris/NaCl/Triton: (aka TBNT) 50 ml 1M Tris pH 7.5 15 ml 5M NaCl 5 g BSA 1.5 ml Triton-100 433.5 ml autoclaved dH2O (up to 500ml) Tris/NaCl/MgCl2 20 ml Tris (pH 9.5) CH in situ protocol January 2011 8 4 ml 5M NaCl 5ml 2M MgCl2 171 ml autoclaved dH2O (up to 200ml) 3. Move slides into box containing 1x PBS (Use PBS box # 3 from day one, with most of the PBS replaced using the remainder of the 2L made on day 1). Leave at room temperature for 5 minutes. 4. Place slides face up in Tupperware box and cover with Blocking Solution (75100ml). Shake gently at room temp for 45 minutes. 5. Pour off blocking solution and replace with 1.0% BSA solution (TBNT). Shake at room temp for 45 minutes. 6. In a tube, dilute anti-DIG antibody 1:1250 by adding 8 l to 10 ml of BSA solution (TBNT) 7. Drain blocking solution from Tupperware, remove slides with the help of a spatula, and place on paper towels. Rinse Tupperware, line with paper towels, wet with milliQ water, and set up pipets to rest slides on. 8. Pour antibody/BSA solution mix into a weigh boat 9. Sandwich two slides together, facing each other (Note, the probe-on slides we are using have raised corners to create space between slides when sandwiched). 10. Place the end of slide sandwich in the weigh boat containing antibody solution. The solution should be sucked up by capillary action. Next, drain this liquid by still holding slides together and resting edge on a paper towel. 11. Repeat capillary action step with slides, but this time do not drain off solution. Rest slide sandwich on pipets in Tupperware box. Cover box and leave at room temperature for 2 hours. 12. Drain slides by resting end on paper towel, then separate them using a razor blade. Temporarily rest slides on paper towels, remove paper towels and pipets and rinse Tupperware. 13. Replace slides face up in Tupperware, add 100ml of BSA solution (TBNT) shake with shaker for 15 minutes at room temp. 14. Drain BSA solution and repeat wash three more times. (4 washes total, each is 15 minutes) 15. Next, wash for 10 minutes with 100ml Tris/NaCl/MgCl2 shaking at room temperature. (Note: Bob Franks does 2 x 15 min w/o shaking, sometimes 3x15 min) CH in situ protocol January 2011 9 16. Temporarily place slides on paper towels, and set up wet paper towels and pipets to rest slides in box. 17. Pour a few mls of Western Blue into a weigh boat 18. Rinse slides by dipping in 50 ml tube containing Tris/NaCl/MgCl2, 19. Sandwich two slides together (facing each other, with white corner feet lined up) and suck up western blue solution using capillary action. 20. Rest sandwiched slides on pipets in Tupperware. Carefully wrap box in foil (Western Blue is light sensitive) and leave in dark cabinet at room temperature over night (and up to 24 + hrs, depending on amount/quality of probe, and expression pattern) Day 3 Protocol: 1. Check the development of slides starting at about 16 or 18 hrs and every few hours afterwards if not done. If you want slides to go longer after separating them for viewing under microscope, repeat capillary action step with western blue and return to Tupperware in the dark. 2. When ready to permanently stop reaction development, separate slides, rinse in TE (pour some into a box or tube) and then air-dry. 3. When dry, mount a coverslip by adding ~3 drops of crystal mount to slides and coverslip. Do not do an ethanol series of dehydrations because western blue is soluble in ethanol and this would cause the signal to be greatly reduced. Trouble shooting: If background is high, less probe can be used, temperature of hybridization can be increased, and western blue incubation time can be decreased. 10X PBS 75.9 g NaCl 18.76g Na2HPO4 (dibasic) 4.1g NaH2PO4 pH to 7, bring up to 1L with dH2O, then DEPC treat overnight and autoclave 20X SSC 175.3g NaCl (3M) 88.23g NaCitrate (300mM) pH to ~ 7.0 with 5 or 10 M NaOH CH in situ protocol January 2011 10 Bring up to 1L with dH2O, DEPC treat overnight, then autoclave 50x Denhardt’s solution Make less than this recipe. Only need 200l for every 8 ml hyb solution 5g Ficoll 400 5g Polyvinylpyrrolidone 5g BSA Bring up to 500ml with dH2O, sterilize or autoclave???