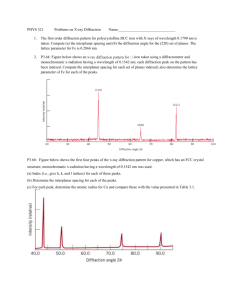
operating instructions siemens powder diffractometer
advertisement

Practical: whole pattern fitting structure (Rietveld) refinement In this practical we will learn hoe to extract information about unit-cell lattice parameters and structural parameters from a X-ray diffraction pattern. To this end we will use the Rietveld method which consist of least square refinements to obtain the best fit between the ENTIRE observed diffraction pattern and a calculated pattern from a simultaneously refined structural model, instrumental factors and background parameters. Diffraction patterns are nowadays collected in digitized form, i.e. as numerical intensity values, yi, at every step increment, i. The increments depends on the method used and can be either in scattering angle, 2, or in some energy parameter as velocity (time-of-flight neutron data) or wavelength (energy dispersive data). For this practical the diffraction patterns have been collected at a fixed wavelength and therefore are given as a function of 2. The first exercise involves structure refinement of a diffraction pattern of Fe-periclase collected at BGI using a Philips X’Pert Pro X-ray diffraction system operating in reflection mode, with CoK1 ( = 1.78897 Å) radiation selected with a focusing monochromator and a Philips X’celerator detector. The sample was mixed with some Si (with less than 2:1 sample:Si ratio) which is used as an internal standard. The second exercise (optional) is more complex, since it involves the unit-cell lattice refinements of diffraction patterns collected at different pressure. The major challenge of such refinements is to get a proper description of the background function. SIMPLIFIED INSTRUCTIONS FOR THE GSAS SOFTWARE. GSAS is a suite of DOS executable programs derived from original Fortran code written for Vax systems. This code was subsequently ported to Unix and Dec/Alpha systems, and then to MSDOS. GSAS is available free, without license. To help support the continued development of GSAS you should provide appropriate acknowledgement in published work (A.C. Larson and R.B. von Dreele, "General Structure Analysis System (GSAS)", Los Alamos National Laboratory Report LAUR 86-748, 2004). GSAS can be used by running the DOS programs directly from a DOS prompt. Input to the programs is then a command-line interpreter. The alternative is to use one of the GUI’s developed for Windows. These GUIs have the advantage that they transfer the necessary parameters between the various programs comprising GSAS, and many provide an editable display of the most important parameters in the refinement. The GUI we will use is the EXPGUI: A GUI developed by Brian Toby (NIST) (B.H.Toby, EXPGUI, a graphical user interface for GSAS, Journal of Applied Crystallography, 34, 210-221, 2001). For this practical you need to install both GSAS and EXPGUI. These programs can be downloaded from http:/www.ccp14.ac.uk/ DATA ANALYSIS To run GSAS you need a measured diffraction pattern and an instrument parameter file which contain detail of the diffractometer such as radiation wavelength geometry and default peak- profile parameters. For the first example the diffraction pattern file is EX1.GSA and the parameter file is PHILIPS.PRM. Note: it is advisable to copy both instrument file and the file with your diffraction pattern into the same folder where you will run the GSAS program. Also keep the name of your folder short and with no space, to avoid problems. - Start EXPGUI (windows shell for GSAS) - Select your directory - Type the name of experimental file without extension and press READ. As the file does not exist yet the File Open Error window appears with the message “file yourfile.exp does not exist in C:/your path. OK to create?” You should select CREATE. - In the next window you are asked to input the title for you experiment. Type a short description of experiment, e.g. exercise 1, then press SET. - The EXPGUI window will appear In order to proceed with the refinement of your data, you need to set up the starting model, i.e. you need to insert into the program the structural parameters of the phases present in your diffraction pattern. In the case of the first exercise the two phases are as already mentioned Fepericlase and the Si standard. To insert information about the phases do the following: - Select the inset Phase and press the Add Phase button. You should see the following window appear. - We input first periclase: type the phase title: periclase, space group: F m -3 m, and a, b, c, α, β, γ: 4.26 and 90° (since it is cubic all axes are the same, but you need still to insert the values in all three fields). Then press Add. A window with the symmetry elements appears. Press Continue. Now you should add the atoms: press the Add new atoms button. The dialog box “Add new atom” appears. - - - If you have more then one atom press the More atom boxes button. The structure of periclase can be described with two atoms, one Mg at x = 0 y = 0 and z = 0 and one O at x = 0.5 y = 0.5 and z = 0.5. For each atom add the atom type (as in periodic table of elements), the atom name (free text, you can leave the default), the atomic positions. Leave the default values for the occupancy, and the Uiso. Then press the Add Atoms button of the dialog box. If your diffraction pattern have more then one phase, repeat the“Add Phase” procedure for every phase present. Repeat the procedure for the Si standard. Space group F d -3 m, lattice parameters a = b = c = 5.43088 (note: you should input all the digits because this is our standard and it will be used to refine the 2 zero of the diffraction pattern), = = = 90°. The structure has one atom: Si at x = 0.125 y = 0.125 and z = 0.125. Change the Uiso value to 0.01, since the atomic vibration of Si is very small. At this point you have your structural model ready, now you need to insert in the program the actual data which need to be refined and the experimental set up. This is done in the “Histogram” inset: - From the main EXPGUI window select the inset Histogram and press the Add new Histogram button. The following window will appear. - In the dialog box select your data file: EX1.GSA and the instrument parameter file PHILIPS.PRM and check that the usable data limit (i.e. 2-Theta Max) correspont to the end of your 2 range which in the case of exercise1 is 100°. Press the Add button on the bottom left corner of the dialog box. Since we have collected the diffraction pattern of two different phases in different amounts we need to set "scaling factors" to be refined which can scale opportunely the two structural models. These are important for quantitative analyses of multi-phase mixtures. - Go to the inset Scaling, uncheck the box for refining the Scale Factor (which would use an unique scale factor for both phases) and check instead the boxes for Phase 1 and Phase 2 to allow to refine the Phase Fractions. The inset should than look like the following: Because the majority of the intensity information in the entire diffraction pattern belongs to the background is it quite important to have a pretty good model for it before actually starting to refine the structural and instrumental model. First of all, we need to chose an appropriate function to fit the background. This is not difficult for well crystalline materials collected with a conventional diffractometer as in the case of this first exercise. The choice of the background is done in the Histogram inset as following: - From the main EXPGUI window select the inset Histogram and press Edit background. A dialog box will appear in which you can chose the function type you wish for describing your background as well as the number of parameters. The most used background functions are Function 1 (Shifted Chebyschev) and Function 2 (Cosine Fourier series). For a detailed description of these functions please refer to the GSAS manual. As a start just use the default background function. We can now start with the first refinement cycles. At this point the only think you will refine are the phase fractions and the background coefficients. - - - - Run the powpref procedure (under the main menu line in EXPGUI window). This set up the least-squares calculation. Run genles and look at the refinement statistics provided in the DOS emulation window i.e. CHI**2 and F(R**2) (Chi square and R-factor). The goal is to minimize these parameters. Since we are just refining the background these statistic parameters will be pretty large. Run liveplot to see the results of your first cycles of refinements. In the liveplot window go to menu file -> Tickmarks and select phases for which you want to see the calculated reflection position. Now you will see your experimental data, calculated diffraction pattern, calculated reflection positions and difference curve. You can zoom in just by selecting the range with your mouse (first click to upper left corner of the box you want to select, second click the bottom right corner. To zoom out use the right mouse click). After every change run POWPREF and GENLES Since the diffraction pattern was collected using a monochromator, it is very likely that your zero position is not correct. So if you are happy with the fitting of your background, refine the zero parameter by checking the corresponding box in the Histogram inset. If you are not happy with the fitting of your background, add more coefficient to the function you are using. NOTE: since Si is a minor phase it is better to refine the zero only for the Si phase without considering the periclase phase. In order to do that uncheck the box of phase 1 in the Histogram inset. Your EXP window should look like this: Once your Si phase is well refined check the box for phase 1 again. NOTE: never refine the lattice parameter of Si since they are used to calibrate your zero!! Step by step select the parameters you want to refine by checking the correspondent box. The usual order is: the zero position (in the Histogram inset), than the unit-cell of your phase (in the Phase inset) and finally the Lorentzian (LY) and Gaussian (GW) profile width (in the Profile inset). The CHI**2 should decrease at every step. The lattice parameters of the periclase should increase with respect to the values you have inserted. This is because the sample is not pure MgO but contain some Fe. Moreover the intensities of some reflections are not properly reproduced by the model. Since the CHI**2 is already quite good (normally a value below 8) we can start to refine the structural model. - In the inset Phase 1 click Add new atom. Add Fe at x = 0 y = 0 and z = 0, and give an occupancy value smaller than 1 (i.e. 0.5). Also change the occupancy of Mg to a value = 1 - Fe occupancy. You can do that by selecting the Mg atom in Phase inset. - Run a cycle of refinements and then select first the Mg and then the Fe atom and check the Refinement flags F to refine their occupancies. Since the two occupancy are not independent because their sum should be equal to 1 we will add a constraint for refining them accordingly. To do that go to the Constraints inset and chose New Constraint. In the dialog box that appears chose Phase 1, select the Mg atom and chose the FRAC variable as shown below - Select New Column and this time again for phase 1 select the Fe atom and give as multiplier -1.0. Then press Save. Run more refinement cycles. At the end you should have reach a value of CHI**2 close to 1 which will indicate that your refined model is very close to the experimental pattern. Background Reading and References If you are unfamiliar with Rietveld refinement, you are strongly advised to obtain at least one of the following two references “The Rietveld Method”, edited by R.A. Young. An excellent introductory guide to pattern fitting. “Rietveld refinement guidelines” McCusker et al. J. Appl. Cryst. (1999) 32:36-50. Another good beginners guide. The following two papers provide some essential background for understanding some of the more subtle issues involved in fitting X-ray powder diffraction patterns: Jansen et al. (1994) J. Appl. Cryst. 27:492-496. (Good summary of how people fiddle refinement parameters). Finger et al. (1994) J. Appl. Cryst. 27:892-900. (The definitive peak profile paper).