Lab Exercise 10 – Transformation of Bacterial DNA
advertisement

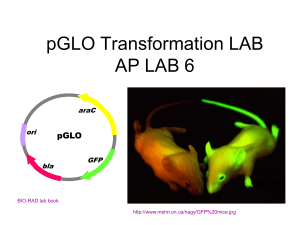
Lab Exercise 10 – Transformation of Bacterial DNA Lab Exercise 10 – Transformation of Bacterial DNA 1. 2. 3. 4. Objectives: Following this exercise the student should be able to: compare and contrast bacterial mechanisms of genetic variation describe the lab methods used to bioengineer traits perform a bacterial transformation evaluate the role of bacterial transformation and use of DNA to modify organisms in light current information on the biotech industry, biological warfare, and medical applications. Background DNA represents the blueprint (genotype) for the physical attributes (phenotype) expressed in organisms and their offspring. Bacteria reproduce via binary fission unlike Eukaryotic cells, which undergo mitosis. Binary fission produces identical daughter cells, or clones. This contrasts with the products of meiosis that produce genetic variability and offspring with completely new combinations of genes. Genetic changes in bacteria changes are often associated with factors that increase pathogenicity by equipping the bacteria with additional abilities to produce toxins, evade the immune system, or resist antibiotics. While eukaryotic cells have most of their genetic material in chromosomes within the nucleus and a small amount in organelles such as the mitochondria, bacterial DNA exists in the nucleoid as a single circular chromosome and as small circular extra-chromosomal DNA called plasmids. Plasmids are not necessary for bacteria to survive. They reproduce at a rate different from the chromosome, and can be transferred to other bacteria altering their phenotype and abilities. Genetic variation in bacteria takes place through three methods: conjugation, transformation, and transduction. In this laboratory exercise you will perform a bacterial transformation. Before looking into the procedure, fill out the chart below to describe the three mechanisms for genetic variation in bacteria. Type of mechanism for Description of factors necessary for this genetic variation to occur Conjugation Transformation Transduction 2 http://www.animalnetwork.com/fish2/aqfm/1999/dec/wb/wbfig6.asp Extract jellyfish GFP gene Create a plasmid with an Ampicillin resistance gene and activator pGLO Ampicillin resistance pGLO plasmid with arabinose activator Insert the plasmid into E.coli K-12 icbxs.ethz.ch/members/ leu/gfp_home.html In this lab you will use biotechnology to transform bacteria (alter their phenotype) using a gene isolated from jellyfish. The gene, called GFP (Green Fluorescent Protein), is naturally found in a bioluminescent jellyfish called Aequorea victoria. The GFP gene has been removed and used to create a plasmid called pGLO. The plasmid will be inserted into E.coli transforming the bacterial genome resulting in bacteria with the capacity to produce GFP. In addition to coding for the fluorescent protein the plasmid also codes for resistance to ampicillin and has a gene-regulating mechanism that switches the gene on in the presence of arabinose (a sugar). A special strain of E.coli is being used called E.coli K-12. This strain does not produce any toxins and can not survive outside of the lab. The transformed cells are selected from the parent cells by culturing on agar made with ampicillin. The susceptible parent cells will not survive, but the new transformed cells are resistant to ampicillin and will grow. The transformed cells grown on media with arabinose will switch the GFP gene on. The procedure will require three main steps. In step one the transformation fluid (CaCl2) neutralizes the phosphate charges in the phospholipid cell membrane and DNA backbone allowing the plasmid (pGLO DNA) to enter the cell. In the second step you will heat shock the cell which further enhances DNA uptake but is highly time sensitive. In the third step the bacteria will be incubated with the appropriate ingredients to: a) select the transformed strain and b) allow transformed cells to express the trait. Materials: Ampicillin resistant glowing E. coli K-12 stock cultures of E.coli K-12 plasmid pGLO transformation fluid (CaCl2) LB broth 5 pipettes loops and incinerators 2 lab groups work together – 6 people each setup 1 LB (Luria & Bertani) plate 2 LB plates with ampicillin 1 LB plates with amp & arabinose microtubes tube holders stop watch ice bath water bath at 42ºC Gram stain equipment and microscope Lab Exercise 10 – Transformation of Bacterial DNA Procedures: 1. Read 230-234 in the text before coming to the lab. 2. Label one microtube +pGLO and another –pGLO. Place them in the microtube rack. 3. Using a sterile pipette transfer 250µl of transformation fluid (CaCl2) into both the +pGLO and –pGLO microtubes. 4. Place the tubes on ice. 5. Using the loop and aseptic technique, pick one isolated colony from the stock plate and place it in the +pGLO microtube. Use the loop to gently mix. 6. Using the loop and aseptic technique, pick one isolated colony from the stock plate and place it in the -pGLO microtube. Use the loop to gently mix. 7. Use the sterilized loop to add a loop of the pGLO plasmid to the +pGLO microtube ONLY. Use the loop to stir the plasmid and stock culture. DO NOT ADD anything to the –pGLO microtube. 8. Incubate the tubes on the ice for 10 minutes. 9. While the tubes are incubating, label the LB plates as follows LB plate: -pGLO: LB LB/amp plate: +pGLO:LB/amp LB/amp plate: -pGLO:LB/amp LB/amp/ara plate: +pGLO:LB/amp/ara 10. Heat Shock: Using the foam microtube holder, transfer the +pGLO and –pGLO microtubes into the 42ºC water bath for exactly 50 seconds. Make sure to push the tubes all the way down in the rack so that they make contact with the water. 11. After 50 seconds, rapidly transfer the microtubes back to the ice bath for 2 minutes. 12. After the 2 minute ice incubation, using a new sterile pipette for each tube, place 250µl of LB nutrient broth in each microtube. Close the tubes. 13. Incubate the tubes 10 minutes at room temperature. 14. Tap the closed tubes with your fingers to mix the contents, then using a new sterile pipette for each tube, pipette Experimental Plates 100µl of the +pGLO tube to the LB/amp plate: +pGLO:LB/amp 100µl of the +pGLO tube to the LB/amp plate: +pGLO: LB/amp/ara Control Plates 100µl of the -pGLO tube to the LB/amp plate: -pGLO:LB/amp 100µl of the -pGLO tube to the LB/amp plate: -pGLO:LB 15. Using aseptic technique, streak the suspensions on the plates. Be sure to incinerate between plates. 16. Stack your plates tape them together and label them with your group name and AM or PM lab. Incubate them, upside down, in the 37ºC incubator. 4 Exercise 10 – Transformation Lab Report Name_________________ Control Plates 1. Observe the plates & count the colonies. Then observe the colonies under UV light. Record the results of your experiment in the table below. Plate Approximate Observations under UV light Number of colonies -pGLO:LB -pGLO:LB/amp Transformation Plates +pGLO:LB/amp +pGLO:LB/amp/ara 2. Under regular light describe any difference between the various colonies? 3. What do we call bacteria of the same species that have different genetic abilities but are still biochemically the same or very similar? (circle all that apply) a. a new species b. a different genus c. a strain of the original species d. a mutant organism e. a transformed organism 4. Ask the instructor if you should Gram stain all four plates and note any differences in the Gram stain. 5. What was the purpose of: the ice bath the 42ºC water baths the transformation fluid (CaCl2) 6. Why was the ampicillin resistance gene added to the plasmid? 7. Why was the activator gene added to the plasmid? Last Updated 2/12/2016 ©Janet Fulks