Assignment 1: Locating sequences using Stringsearch and Mapping
advertisement

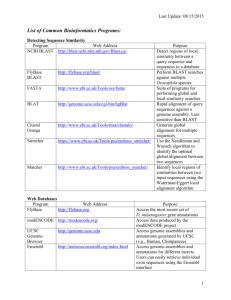
Assignment 1: In this assignment you should prepare a report with the answers to various questions. This report can be written in any text editor or word processor. Add your name and e-mail address on the front page!!! Part 1: Locating sequences with the ENTREZ: http://www4.ncbi.nlm.nih.gov/entrez Can you find a sigma factor in any mycoplasma? Repeat the same task for bacillus. Please write the following in your report: 1. How many sequences were searched (separately for mycoplasma and bacillus)? 2. How many hits were reported? 3. How many representatives of mycoplasma (bacillus) are completely sequenced? 4. Do you think that you have found all sigma factors in all mycoplasmas? Choose one of the sequences to be viewed. Present the data of the chosen sequence Download the chosen sequence into your current directory type: Please write the following in your report: 1. The sequence code (full name)? 2. The sequence accession number? 3. The sequence description? 4. The sequence length? DNA Sequence Analysis using fuzznuc Mapping restriction enzyme cleavage sites using restrict Locating sequences with the fuzznuc program http://bioweb.pasteur.fr/intro-uk.html#dna 1. Can you find any complete bacterial genome, in which you can map patterns (for retrieval a bacterial genome use ftp protocol and save *.fna file) a. TTTTTxxTTTT b. TATATATATATA with two mismatches c. CACATATGTG 2. Select three different restrict enzymes and map them on both strains of Agrobacterium tumefaciens complete genome. Compare results. Please write the following in your report: 1. How many hits were reported? 2. Present the results in the attractive format – see example from the previous semester. Assignment 1 Moshe Cohen (Mussa) e-mail: mch@yahoo.com Part 1. 1. 23180319195 sequences were searched. 2. 3208 hits of fibroblast growth factor were reported. The chosen sequence is: Homo sapience fibroblast growth factor 11(FGF11) gene; accession number AY094623 version gi20160214; sequence length is 6447 bp. Part 2. 1. 8 whole bacterial genomes were searched, only in blablabla genome the patterns could be mapped: a) TTTTTxxTTTT – ?? matches. b) TATATATATATA – 344 matches: 126 patterns with 0 mismatches; 44 patterns with 1 mismatch; 174 patterns with 2 mismatches. c) CACATATGTG – 10 matches. 2. Restriction enzyme cutting sites mapping on the complete genome of Streptococcus pyogenes, strain M1 GAS Restriction enzyme Hae III EcoRII MboI Number of cuts 4 gg/cc 3 /ccwgg 4 /gatc Position of the cuts 189,378,566,712 224,408,1022 270,464,786,939