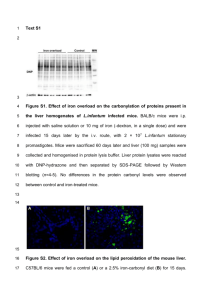
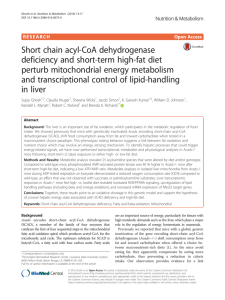
hep22363-SupplementaryMaterialsandMethods
advertisement

Materials and methods Online Mice and diet. Mice were housed under standard conditions given free access to food and water. Experiments were performed according to Dutch laws, approved by the Committee for Animal Welfare of Maastricht University. Several experiments were performed. In the first experiment, 12-week old female C57BL6 and ldlr-/- mice were fed a high fat diet containing 21% milk butter and 0.2% cholesterol (HFC), for 2, 4, 7 and 21 days. A single group was kept on a standard chow diet until the age of 12 weeks and served as control group. In the second experiment, male C57BL6, ldlr-/- and APOE2ki mice were put either on HFC for 7 days or were kept on chow diet. In the third experiment, male and female C57BL6, ldlr-/-, and APOE2ki mice were fed HFC or the high fat diet without the added cholesterol (HFnC) for 7 days. The HFnC diet contained only the residual cholesterol derived from the butter component, i.e. 0.05%). Collection of blood, sacrificing of the mice, and tissue isolation was done as described previously (8). Lipid analysis Approximately 50 mg of frozen liver tissue was homogenized as described previously (8). Both plasma and liver lipid levels were measured with enzymatic color tests (1489232, cholesterol CHOD-PAP, Roche, Basel,Switzerland; TR0100, TG GPO-trinder, Sigma Aldrich, St. Louis, MO, USA; 999-75406, NEFAC, ACS-ACOD, Wako Chemicals, Neuss, Germany) as described before (8). RNA isolation and first strand cDNA synthesis Total RNA was isolated from approximately 25 mg of mouse liver tissues as described previously (8). All applications were done according to manufacturers protocols. Total RNA (500ng) from each individual mouse was converted into first strand cDNA with iScript cDNA synthesis kit (170-8891, Bio-Rad, Hercules, CA, USA) according to the manufacturers instructions. Quantitative PCR The changes in gene expression of inflammatory markers were determined by quantitative PCR on a Bio-Rad MyIQ with the IQ5 v2 software (Bio-Rad, Hercules, CA, USA) by using IQ SYBR Green Supermix with fluorescein (170-5006CUST, Bio-Rad, Hercules, CA, USA) and 10 ng of cDNA. For each gene a standard curve was generated with a serial dilution of a liver cDNA pool. To standardize for the amount of cDNA, Cyclophillin A (Ppia) was used as the reference gene. Primer sets for the selected genes were developed with Primer Express version 1.5 (Applied Biosystems, Foster City, CA, USA) using default settings. Primer sequences: MCP1-forward, 5’-GCTGGAGAGCTACAAGAGGATCA-3’; MCP1-reverse, 5’-ACAGACCTCTCTCTTGAGCTTGGT-3’; CD68-forward, 5’-TGACCTGCTCTCTCTAAGGCTACA-3’; CD68-reverse, 5-TCACGGTTGCAAGAGAAACATG -3’; TNFa-forward, 5’-CATCTTCTCAAAATTCGAGTGACAA-3’; TNFa-reverse, 5’-TGGGAGTAGACAAGGTACAACCC-3’; Ppia-forward, 5’-TTCCTCCTTTCACAGAATTATTCCA-3’; Ppia-reverse, 5’-CCGCCAGTGCCATTATGG-3’. Data from qPCR was analyzed according to the relative standard curve method. Taqman® Low Density Arrays To perform gene-expression analysis on a medium scale, Taqman® Low Density Arrays (TLDA) 96a (PN 4342259) were used. Each TLDA plate contained 4 x 96 annotated and validated individual TaqMan® Gene Expression Assays (forward primer, reversed primer and Taqman® Probe) (supplementary table 1). Per individual assay 2ng cDNA of a single liver was loaded together with the TaqMan® Universal PCR Master Mix (PN 4324018). Of each group, mRNA from 5 individual mice was used. TLDA plates were run on an ABI Prism 7900HT Sequence Detection System with a TaqMan® Low Density Array Upgrade. Data was analyzed by using RQ Manager 1.2 software. Technically failed assays were omitted from analysis. Materials, equipment and software necessary to perform TLDA gene expression studies were obtained from Applied Biosystems, Foster City, CA, USA. All data was normalized to Ppia expression. Significant differences were determined with student t-tests; p-values < 0.05 were considered significant. Liver histology 4µm paraffin embedded liver sections were stained with Haematoxillin/Eosin (HE) and Periodic acid Schiff (PAS)-diastase. Frozen liver sections (7µm) were fixed in acetone and stained with CD68 (FA11) or Mac1 (M1/70). Pictures were taken with a Nikon digital camera DMX1200 and ACT-1 v2.63 software (Nikon Corporation, Tokyo, Japan). Electron microscopy. Livers were freshly isolated from the mice and perfused and fixed overnight with 2.5% glutaraldehyde (Ted Pella, Redding, CA, USA). Tissue fragments were washed and post fixed in 1% osmium tetroxide. Tissues were subsequently dehydrated trough 100% ethanol, cleared with propylene oxide, and embedded in epoxy resin. Sections of 1 μm were stained with toluidine blue to identify the presence of foamy Kupffer cells. Next, sections of 70-90 nm were cut on an ultra-microtome, mounted on Formvar-coated (1595E, Merck) 75 mesh copper grids and counterstained with uranyl acetate and lead citrate before analysis on a Philips CM100 transmission electron microscope. Statistical analysis Data were analyzed using Graphpad Prism 4.0. Groups were compared using 2-tailed nonpaired t-tests or ANOVA with a Dunnet post test, based on the statistical relevance. Data is expressed as means SEM and considered significant at p 0.05.