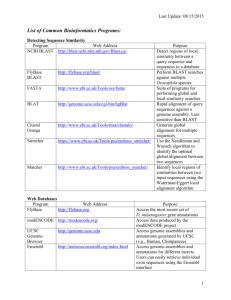
Genomic Science Facilities and Resources
advertisement

GENOMIC SCIENCES FACILITIES & RESOURCES Buildings Training program faculty laboratories are located in a cluster of buildings on Science Hill at the UCSC campus. The Jack Baskin Engineering Building (JBEB), constructed in 1971, together with the Engineering 2 (E2) Building, completed in 2004, provide 200,000 square feet of teaching, research, and office space for training program faculty in Biomolecular Engineering, Applied Mathematics and Statistics, and Computer Engineering. The Electrical Engineering, Computer Science, and Technology and Information Management departments are also housed in these buildings. Built in 1989, the Sinsheimer Laboratories building provides 98,000 square feet of space for approximately 32 laboratories and associated meeting and office space. It is home to investigators from the departments of Molecular, Cell, and Developmental Biology, Chemistry and Biochemistry, and Biomolecular Engineering. Designed around an open central stairwell and atrium, the building maximizes contact between faculty, students, postdocs, and research staff. The Biomedical Sciences Building (Biomed), completed in 2011, is designed to promote interdisciplinary interactions and houses faculty from the departments of Molecular, Cell, and Developmental Biology, Chemistry and Biochemistry, Biomolecular Engineering, and Microbiology and Environmental Toxicology. The building provides approximately 92,000 square feet of space for approximately 20 laboratories and facilities, including a state-of-the-art vivarium for animal research, which augments an existing vivarium located next to Sinsheimer Laboratories. The Physical Sciences Building (PSB) houses investigators from the departments of Chemistry and Biochemistry, Microbiology and Environmental Toxicology, and Biomolecular Engineering. Completed in 2006, it is designed to foster interaction among researchers exploring human and environmental health. This five-story building has 132,000 square feet state-of-the-art lab, classroom, and support facilities to serve a diverse group of scientists. Faculty Laboratories The research laboratories of our program faculty range in size from 1,200 to more than 5,000 square feet and are well equipped to perform the projects offered to our trainees. All labs have access to support facilities including shared equipment rooms, autoclaves, dishwashing, controlled temperature rooms, fume hoods, cell culture facilities, etc. Faculty laboratories also include computational research space in cubicle-partitioned rooms and offices located in E2, PSB, and Biomed to provide workspace for graduate students, postdoctoral scholars, research and technical staff and visiting scholars. Computers are either Dell desktop workstations or IBM, Apple, or Dell laptops; some staff have both desktop setups and laptops. Center for Biomolecular Science and Engineering (CBSE) Resources Major Computing Equipment: Located in JBEB room 213: 256 quad core Intel Xeon compute nodes (npk cluster, 8 GB memory each, 1024 cores total) 2 computers (500 GB memory and 64 processing cores each, 20 TB local disk space) for high-availability virtual machine hosting 8 file servers (nest filesystem, 80 terabytes of replicated networked data storage total) 6 backup servers with capacity to store 660 terabytes of data Located in E2 5th floor: 49 desktop workstations and 27 laptops for staff, postdoctoral researchers, graduate students, telecommuters, and visitors Located in UCSC ITS Data Center: 8 dual 12-core AMD processors (RR, 6 x 128 GB, 2 x 64 GB memory) for web serving of genome browser 4 BLAT servers (128 GB memory each) 3 file servers to centralize the data served by the genome browser (150 TB of data) Download server (100 TB available storage) Located at San Diego Supercomputer Center: 32 dual 16-core AMD compute nodes (ku cluster, 256 GB memory each, 1024 cores total) 68 dual 16-core AMD compute nodes (podk cluster, 256 GB memory each, 2176 cores total) 3 large memory computers (1 TB memory and 64 processing cores each, 24 TB local disk space) for software & database development 16 file servers (hive filesystem, 12 data servers, 4 metadata servers, 600 terabytes of replicated networked data storage total) 1 Hitachi HUS-VM file storage system with 1 PB usable networked storage (pod filesystem) Redundant/Load-balancing download and networked file server (100 TB available storage) 2 file servers dedicated to ENCODE data (100 TB available storage each) Computational clusters: To support UCSC's Genome Browsers and associated tools, databases, and genome research, the Center for Biomolecular Science and Engineering (CBSE) runs three parallel processor facilities, currently managed and supported by 3 FTE system administrators. The newest of these, ku, is comprised of 8 Supermicro units, each with 4 blade servers, and each blade server has 2 x 16-core AMD processors, 256 gigabytes of memory and 2 terabyte of disk space. That gives us a total of 1024 compute cores, 8 terabytes of memory and 64 terabytes of local storage to the cluster. The npk cluster consists of 256 quad core Intel Xeon compute nodes, each with 8 gigabytes of memory, totaling 1024 cores and running on a Linux operating system. We also installed a new cluster behind a firewall to be used for computation on restricted access data. This cluster, podk, consists of 17 Supermicro units, each with 4 blade servers, and each blade server has 2 x 16-core AMD processors, 256 gigabytes of memory and 4 terabyte of disk space. The grand total for the podk cluster is 2176 cores, 17 terabytes of memory and 272 terabytes of local storage. All these systems were designed to provide an exceptional amount of inexpensive computing power in minimal space. Centralized data: The ku cluster and development servers are supported by a group of file servers (12 data servers and 4 metadata servers running IBM GPFS), providing almost 600 terabytes of usable, replicated network storage. The cluster is connected with 10-gigabit ethernet to our network infrastructure. This data is backed up to our offsite backup servers, with current capacity of 660 terabytes. Behind the firewall, we have a Hitachi fileserver that has over 1 petabyte of available storage. We also maintain the UCSC Cancer Genomics Hub (CGHub), which has deployed a computing environment to serve the purpose of archiving and distributing TCGA cancer data to participating labs and researchers around the world. The environment includes a GPFS distributed filesystem that is 4.8 petabytes large, along with four 10Gb/s connected firewalls that protect the data inbound and outbound. There is a cluster behind the firewalls comprised of sixteen application servers that process incoming data and also serve the data to authorized users worldwide. Network infrastructure: We currently have a Cisco based network backbone, with the entire backbone being linked at 10Gb/s. In the San Diego Supercomputer Center, all our machines are internally connected using a 10Gb/s infrastructure provided by Cisco Nexus 3064PQ switches. In our main UCSC datacenter, we utilize a Cisco 6509-E and and Cisco 6506-E for private and firewalled area switching, as well as several 3750-E switches for edge switching to the UCSC core. We have about a dozen end-hosts connected at 10Gb/s as well to the 6506-E for high speed access to our firewalled space as well as our GPFS filesystem and cluster environment. The internal backbone in our main datacenter extends to two satellite locations in shared datacenters around campus, in which we have a small presence. These satellite locations are connected at 10Gb/s through 3750-E switches. Development computers: CBSE employs 3 main computers with 64 computing cores and 1TB of memory each for software and database development. These development servers attach to 24 TB of local disk space. We also provide a high-availability connected computer setup for virtual machine hosting. These redundant machines have 64 computing cores, 500 GB of memory, and access to 20 TB of local disk space. Web servers: The web servers for the UCSC Genome Browser consist of 6 dual 12core AMD processors that each offer 1 TB of internal disk storage and 128 gigabytes of memory. These machines have access to a central file server that provides up to 100 extra TB of shared disk area, and a couple of central Mysql database servers that holds up to 50 TB of genomic data. Four additional servers provide web access to BLAT (Blast-like alignment tool) software. Each of these machines has 128 gigabytes of memory, since BLAT is a memory-intensive application. Some of the other services we provide include a genome-preview server that allows the public to access our raw data before it has gone through QA, a public mysql server that hosts all our mysql data, a custom track server to store user-generated custom tracks, and a wiki server that holds public information and can keep track of named sessions. We are also working on providing systems in different geographical areas to reduce the access penalty, and the first European system has been deployed in Bielefeld, Germany. Finally, a local download server allows users to download our data; it serves nearly 2 TB of data every day. We also house 1 identical download server at the UCSD SDSC colocation center for load-balancing and redundancy. All of the machines serving the genome browser data are housed in a data center that is designed to function 24/7, 365 days a year. Specialized Facilities, Core Facilities and Shared Equipment The UCSC Paleogenomics Lab has state-of-the art molecular biology and computational resources. The Molecular Biology and Sequencing lab houses an Illumina MiSeq sequencer, a Covaris bioruptor, and a Pippen Prep Blue automated DNA size selection and collection apparatus. The Ancient DNA Handling Facility, overseen by program faculty Prof. Beth Shapiro, is a recently renovated 750 sf laboratory dedicated to handling degraded sample and ancient DNA. Isolated from other genetics processing/characterization facilities, the ancient DNA handling facility is a dedicated clean room in which the external surface of all equipment/consumables entering the facility are cleaned with 10% bleach solution and UV irradiated. Users of this facility wear sterile shoe covers, body suits, gloves and facemasks, and cannot enter the facility after using a non-sterile facility. The facility receives separate airflow from the remainder of the building and has positive air pressure. In addition to standard ancient DNA extraction and PCR protocols, the ancient DNA clean room is completely equipped for preparing samples for next generation sequencing on an Illumina HiSeq system. Compute resources in the Paleogenomics Lab include a NVIDIA Tesla GPU CUDA-based 12-node cluster, a 32-node Linux cluster, and a large memory machine (512Gb of RAM) for the purpose of read-mapping and reference guided assembly, 100TB fileserver, and 10 linux workstations. In addition, we have access to the compute resources that are part of the Jack Baskin School of Engineering (BSOE), which include a 7-node (304 core) CentOS system, and a virtualized Solaris 10 SPARC system with 8 virtual processors on a Sun T2000 (32 thread 8 core 16GHz). The department of Applied Mathematics and Statistics (AMS) and the Baskin School of Engineering (BSOE) maintain a state-of-the-art computer network to facilitate instruction, research, and departmental administration. AMS provides a computer network which includes Mac OS X, MS Windows clients, and a high performance computational cluster. BSOE provides additional computing resources; computer clusters and UNIX/Linux servers. The school provides a PC laboratory to graduate students. The system is maintained by full-time system administrators. The most important element of the AMS computer operation is the availability of innovative mathematical and instructional software and free computer resources that create an environment conducive to experimentation and exploration by our faculty and students. The Nanopore Experimental Laboratory in JBEB is a 490 sf room equipped with five nanopore stations available for research training. Each station has a vibration isolation table, amplifier, digital to analog converter, temperature regulator, faraday cage, microscope, fiber optic light and custom machined parts for the nanopore devices. The nanopore stations are each equipped with a dedicated computer with all software and hardware necessary to perform and analyze nanopore experiments. Project personnel have full-time access to laboratory desktop computers equipped with Clampfit 10 software for nanopore data analysis, MatLab, LabView, and other standard software. The nanopore stations are automatically backed-up by a NetApp FAS2040, a highly scalable dual-controller RAID based network attached storage device, with 20 Tb of total storage. Located on the basement floor of the Biomedical Sciences Building, the Vivarium has 19,000 sf dedicated to animal research. It is in close proximity to research laboratories that use animals. The facility is equipped to accommodate mice and rats, transgenic and immune suppressed mice, hamsters, and guinea pigs. It is also equipped with a state-of-the-art cage washer, walk-in autoclaves, and a vapor hydrogen peroxide decontamination chamber, micro isolators, cubicles, ventilated cage racks, and anesthesia machines to ensure compliance with all statutory requirements. The vivarium includes a transgenic core facility which houses advanced equipment for the production of transgenic animal models through traditional and advanced embryo manipulation techniques. The Vivarium received its accreditation from the American Association for Accreditation of Laboratory Animal Committee (AAALAC) in July 2008, and has maintained its accreditation since that time. The Genome Technology Center (GTC) in JBEB provides “next-generation” sequencing services via state-of-the-art equipment and technologies. GTC equipment includes: Next-Generation Sequencing Platforms, an Illumina Genome Analyzer HiSeq 2000 sequencer, an Applied Biosystems SOLiD 4 sequencer, a Roche 454 Titanium sequencer, a Biotage BSQ96 Pyrosequencer, and Nanostring nCounter. The Center also houses the following automated systems: Luminex L200, a BioDot Microarray Robot, a NorDiag 8000+ Liquid Handling Robot, Stratagene qPCR, Covaris S2 DNA Ultrasonic DNA Shearing, and HydroShear DNA Shearing equipment. The Center is directed by program faculty Prof Nader Pourmand. UCSC's Chemical Screening Center (CSC) houses high-throughput screening (HTS) robotics that are used to identify biologically active compounds and siRNAs targeted toward a variety of biological systems. Investigators can test up to 30,000 chemical compounds per day for biological function and/or usefulness in fighting diseases, such as cancer, malaria, Parkinson's disease, and cholera. The CSC houses liquid-handling robotics, detectors, imaging equipment, and compound libraries. Equipment includes a Perkin Elmer Janus MDT system, a Perkin Elmer Janus Varispan system, a BioTek plate washer, a Matrix WellMate system, Perkin Elmer EnVision detectors, a Molecular Devices ImageExpress imaging system, and a 55,000 compound commercial collection from ChemDiv, as well as a growing collection of marine natural products from Phil Crews’ and Roger Linington’s research groups at UCSC. The Center is located in PSB and supervised by program faculty Prof. Scott Lokey and managed by specialist Walter Bray. The Microarray Facility is used for genome-wide splicing and expression analyses of diverse organisms, from microbes to humans. It supports both spotted microscope slide and Affymetrix microarray research. Equipment includes an Affymetrix GeneChip system, a robotic microscope slide arrayer, an Axon slide scanner, and a 96-channel automated liquid handler (Hydra, Robbins Scientific). The UCSC Institute for the Biology of Stem Cells (IBSC) Stem Cell Facility in Biomed is an advanced research laboratory for the culture and analysis of stem cells. Manager Bari Nazario offers expertise in experimental design, protocol development, and data analysis. This facility enables work on non-NIH-approved human embryonic stem cell (hESC) lines. The facility includes two core cell BSL2 culture rooms, one of which can be dedicated as needed to viral transductions and other procedures warranting a higher level of containment. The stem cell laboratory houses equipment required for cell culture, for recording images of living cells, and for applying functional genomics techniques to the study of stem cell biology, including a highthroughput assay station consisting of a 96-well format luminometer and fluorometer. The UCSC CIRM Shared Stem Cell Facility (SSCF) in Sinsheimer is an advanced stem cell laboratory for research and training in manipulation techniques and the production of transgenic organisms. It supports faculty-led research and the laboratory course for the UCSC CIRM Training Program in Systems Biology of Stem Cells. This facility enables work on non-NIH approved human embryonic stem cell (hESC) lines. The SSCF suite includes a core cell culture room and a cell culture training room for instructing individual researchers, small groups, and students taking the stem cell biology laboratory course. The Flow Cytometry Facility, located in Biomed, houses a state-of-the-art high-speed cell sorting and analysis system, including a BD Biosciences FACSAria cell sorter and a BD Biosciences LSRII cell analyzer. The facility also provides licensed FlowJo software for data analysis and display. This system allows simultaneous detection of up to 14 parameters; up to 4 cell populations can be sorted simultaneously at high purity and deposited in bulk, or as single cells into a range of standard test tubes, multiwell plates, or slides, allowing convenient and flexible analysis of purified cells. Additional specimen analysis equipment includes a Perkin Elmer luminescence counter, a Molecular Devices SpectraMax Plus high-throughput micro-plate spectrophotometer, Agilent Bioanalyzer 2100, ABI Real-time PCR ViiA 7, and a Diagenode CHIP SX-86 IP-STAR. The Flow facility is managed by Bari Nazario. UCSC's Life Sciences Microscopy Center in the Sinsheimer Laboratories building is a shared-use facility that provides cutting-edge imaging options for advanced biomedical research. It includes a Leica DM5500 B wide-field microscope, a live-cell imaging station with a Zeiss Axiovert 200 inverted microscope and motorized z-stage, a Leica SP5 Spot Scanning Confocal, and a Zeiss LSM5 Spot Scanning Confocal. A professional microscopist, Dr. Ben Abrams, manages the Center and provides assistance to users in instrument operation, sample preparation, and experimental design. Several individual faculty members also maintain systems that are available to investigators. These include a PerkinElmer Volocity Spinning Disk confocal microscope, a Leica SP3 confocal microscope, a Prairie Technologies Ultima-IV two-photon microscope, and a DeltaVision wide-field fluorescent microscope with iterative deconvolution technology. The Electron Microscopy Laboratory houses a JEOL 1230 Transmission Electron Microscope (TEM) equipped with a LaB6 gun, a Gatan Cryotransfer holder, and a Gatan Ultrascan digital camera. This instrument is used primarily for work requiring the cryostage and high resolution camera. Equipment is also available for routine specimen preparation for light and electron microscopy. This includes an AO Ultracut ultramicrotome, a LKB glass knife maker, a SBT Model 590+ Tripod Polisher, and an SBT Model 900 Grinder Polisher. Other equipment useful for general specimen preparation includes an Ultrascience 102S high purity glass still, and a Pella3450 Microwave Tissue Processor. Prof. Melissa Jurica manages the instrument. Life Sciences Microscopy Center Facility Manager Ben Abrams is in charge of day-to-day operation. The Electron Spin Resonance (ESR) Facility houses two instruments for examining the structure and properties of metal-containing inorganic complexes, peptides, proteins, enzymes, nanoparticles, and biological membranes. The facility's Bruker ELEXSYS 580 X-band spectrometer operates in either continuous-wave or pulsed mode, with variable temperature control. The instrument is also capable of excitation at two frequencies, thus enabling Double Electron-Electron Resonance (DEER) for measuring long distances between paramagnetic probes. The second instrument, a high-sensitivity Bruker EMX, is especially useful for the limited sample sizes often encountered in biological studies. This facility is managed by Prof. Glenn Millhauser. The Macromolecular X-ray Crystallography Facility houses high-throughput crystallization robotics, a rotating anode/imaging plate X-ray crystallography data collection suite, a cryosystem, and a collection of Apple, PC and Linux computer workstations and software for crystallographic computations, molecular visualization, and model building. The Mass Spectrometry Facility currently houses two mass spectrometers: a Thermo Finnigan LC/MS/MS (LTQ) and an Ettan MALDI-TOF. This equipment is capable of determining the molecular weight of both small molecules and peptides, identifying proteins, and characterizing protein modifications. The facility is located in PSB and supervised by Prof Ted Holman and managed by Mass Spec Specialist Qiangli Zhang. The Nuclear Magnetic Resonance (NMR) Facility brings together an interdisciplinary group of researchers from the departments of Chemistry and Biochemistry, Microbiology and Environmental Toxicology, Molecular, Cell, and Developmental Biology, and Biomolecular Engineering. Ongoing research includes structural elucidation of anti-cancer and anti-viral natural products isolated from marine organisms, organic intermediates for drug synthesis, specially designed peptide intermediates, and tumor suppressor proteins. At present, the facility manages three high-resolution NMR spectrometers: two 3-channel Varian Unity+ 500s with direct and inverse detection probes and a Varian INOVA 600 system with a triple resonance coldprobe system. Initial funding came from the Lucille P. Markey Charitable Trust and the W. M. Keck Foundation, as well as individual research grants from the National Institutes of Health, the National Science Foundation, and other sources available to UCSC. The facility is supervised by NMR Specialist Dr. Hsiau-Wei Lee. Additional resources provided by the university to support research include a fully supported machine shop and electronics shop for custom fabrication and repair work. Machining can be done by machine shop staff or by trained students in an adjacent student machine shop. Furthermore, approximately two dozen conference and meeting rooms (10–100 seats) are available in the Science Hill laboratory buildings; many have teleconferencing and videoconferencing facilities (webcam or videocam; some with plasma screens); several have built-in video/data projection, sound systems, Mac and PC computers with web and presentation software; 1 has a computercontrolled 4-screen projection system. Finally, as one of the three UC campuses that comprise the California Institute for Quantitative Biosciences (QB3), UCSC faculty and their laboratories have direct access to a wide array of core facilities that support genomics and informatics at UC San Francisco and UC Berkeley (http://qb3.org/research/core-facilities#anchor4). UCSC’s Genomic Sciences Environment & Institutional Support At the campus level, UCSC’s commitment to genomic sciences is evidenced by “Genomics and Health” being named as one of the four signature initiatives in the campus’s comprehensive fundraising campaign. With a tagline of “Daring to imagine the defeat of cancer and other devastating diseases,” the campaign is aiming to raise funds to support new PIs, research fellowships, computing infrastructure, and professional program staff. The campaign states that graduate student research fellowships will be vital to UCSC’s current work and will create next generation leadership in the field. Genomic sciences is the primary focus of UCSC’s Center for Biomolecular Science and Engineering (CBSE). Directed by Biomolecular Engineering Professors David Haussler and Joshua Stuart, and also under the scientific leadership of Dr. James Kent, CBSE promotes and supports genomic and stem cell research, technology innovation, and education. An umbrella organization of the Jack Baskin School of Engineering and the Division of Physical & Biological Sciences, the center supports a vast array of biological and engineering research that is fueling biomedical advances and the biotechnology explosion. CBSE’s roots go back to 1985, when UCSC scientists met with a group of international visionaries at a conference hosted by Chancellor Sinsheimer, a meeting that triggered the inception of the Human Genome Project. Fifteen years later, UCSC scientists helped the Human Genome Project reach a stunning milestone by providing the computational solution that produced the first assembly of the human genome, the map of our genetic make-up. Out of these accomplishments and a growing interest in crossdisciplinary science grew the Center for Biomolecular Science & Engineering. The center fosters interdisciplinary research and academic programs that address the scientific questions of the post-genomic era. CBSE Goals Promote interdisciplinary research in areas that encompass the study of genomic information Support the UCSC Genome Browser and the UCSC Cancer Genomics Browser, crucial resources for the international scientific community Support core facilities, such as the computational cluster used for the UCSC Genome Browser and genomic research, the microarray facility, and embryonic stem cell facilities Help meet the need for trained professionals in industry and academia by developing training programs in the areas of bioinformatics and biomolecular engineering Attract research funding for the center, for affiliated faculty, and for students from federal, state, and private agencies Cultivate and maintain mutually beneficial relationships with industry through research collaborations, internship opportunities, and gifting programs. The Biomolecular Engineering (BME) department within the Baskin School of Engineering will serve as the administrative home for the proposed training program and shares CBSE’s primary focus on genomic sciences. The BME department features an interdisciplinary blend of engineering, biology, chemistry, and statistics designed to foster collaboration with other departments. This blend reflects the department’s vision of the direction that biomedical discovery will take over the next two decades. Research groups within the department include UCSC Genome Bioinfomatics, the Systems Biology Group, the Biosensor and Biotechnology Group, the Nanopore Project, Genome 10K, and the UCSC Paleogenomics Lab. Examples of Genomics Projects and Resources at UCSC UCSC develops and hosts the UCSC Genome Browser and the UCSC Cancer Genomics Browser. The Genome Browser, supported and continually developed by approximately 20 scientists, engineers, postdocs and graduate student researchers is used by more 100,000 biomedical researchers worldwide receiving about 3 million page requests per week. The UCSC Cancer Genomics Browser is one of the main data portals for the National Cancer Institute’s (NCI’s) Cancer Genome Atlas (TCGA) project. The Cancer Genomics Browser was developed through the collaboration of several groups and projects in which UCSC participates─the Stand Up To Cancer (SU2C) Breast Cancer Dream Team, the UCSC-Buck Institute Genome Data Analysis Center for TCGA Research Network, and the adaptive I-SPY 1 clinical trial for high risk breast cancer. The Cancer Browser enables teams of researchers to deposit and distribute multidimensional and multiplatform high-throughput datasets and provides the ability to connect to other web-based resources for data analysis including the Broad’s Firehose and MSKCC’s cBio portal. In addition to data visualization and on-line exploration, UCSC also develops state-ofthe-art deep computational tools to investigate the biology of cancer through the analysis of genomic, epigenomic, and functional-genomic datasets on tumor samples. Led by Professor Josh Stuart, the UCSC Systems Biology Group has developed the PARADIGM integrative pathway analysis and related algorithms that are now part of the minimal set of analysis routines to be run on every tumor type for the NCI’s TCGA project. The methods have been used to uncover novel pathways involved in ovarian cancer, connections between Wnt and TGFBeta pathways in colorectal cancer, mechanisms of drug response in triple negative cell line models of breast cancer, and Ki-67 response in luminal breast cancer. We have also developed algorithms that extract reliable information about mutations present in cancer samples from whole genome sequencing, exome sequencing and RNA sequencing, and tested these against similar tools from the Broad Institute, Washington University St. Louis, and Baylor College of Medicine. Other resources developed by the Systems Biology Group include TieDIE, the Tied Diffusion of Interacting Events algorithm for identifying linking networks and pathways between sets of functionally related genes, and The Interaction Browser, a web application for visualizing biological pathways and networks. In the UCSC Genome Technology Center, Professor Pourmand and team use highthroughput sequencing and bioinformatics to understand genetic variation and its function in health, disease, and biological systems. The GTC is aimed at supporting local scientific communities in generating data and resources for basic and applied biomedical research. Other projects in the GTC include technology development to address key problems related to single cell genomics, proteomics and single cell analysis. A nanopipette, developed by Pourmand, allows researchers to study the genomics of single cell (currently they are investigating single neurons) with temporal resolution. The small diameter of the nanopipette provides a non-invasive method for RNA retrieval that extracts just enough material for single cell sequencing without impairing cell function or survival. The Paleogenomics Lab is a co-directed by two training program faculty mentors (Green and Shapiro). Current projects in the lab focus on genomics and population genetics (investigating patterns and processes in genome evolution), ancient DNA (extracting and characterizing DNA from fossil remains) and pathogen evolution (understanding the evolutionary constraints underpinning diversity and evolution in pathogens). The lab is principally focused on developing new experimental technology for recovering DNA from biological remains and computational tools for making biological inferences from these DNA data. NCI has named UCSC a Trusted Partner of the NIH to coordinate the collection of the nation’s cancer genomics datasets through the new portal, the Cancer Genomics Hub (CGHub). CGHub is a secure repository for storing, cataloging, and accessing cancer genome sequences, alignments, and mutation information from the Cancer Genome Atlas (TCGA) consortium and related projects. CGHub gives scientific researchers the statistical power of large cancer genome datasets to attack the molecular complexity of cancer. Projects in Phil Berman’s lab are focused on discovering a vaccine to prevent HIV infection. Current approaches involve ancestral reconstruction to identify HIV envelope sequences giving rise to broadly neutralizing antibodies, as well as full genomic sequencing in an effort to identify polymorphisms in the host that favor the formation of broadly neutralizing antibodies. Both the ancestral reconstruction and human genome sequencing studies depend on collaborations with other members of the BME department including David Haussler, Todd Lowe and Richard Green. Several training program faculty (Haussler, Green, Ares, Lowe, and Sanford) are members of the UCSC RNA Center, which spans several departments and is led by Professor Harry Noller (ribosome structure and function). There is a monthly RNA Club with speakers from both UCSC and from surrounding universities (Stanford, UCSF, Berkeley, Davis), as well as several journal clubs. Institutional Support In addition to the space, facilities, and resources listed above, UCSC will provide several other forms of support for the program. Trainees will be supported by the Graduate Advising office of the Baskin School of Engineering, which helps ensure trainees stay on track with course and program requirements, and helps identify fellowship opportunities and other forms of graduate support. The program will receive accounting and administrative support from the division’s research accountants and department staff, who will help the program run on a daily basis. CBSE grant administrators will provide support for progress reports and other periodic needs of the program. The training program will be further supported by the recruitment and retention resources provided by the CBSE Research Mentoring Institute (RMI), whose director will provide professional development support for trainees and will act as advisor to training faculty on matters of recruitment, retention and mentoring. As mentioned in the letter of support from Scott Brandt, Vice Chancellor, Office of Research, the BME department will provide release time each year to PD Green so that he can dedicate sufficient effort to the program. In addition, campus will also fill any gap in stipend or fee support for program trainees, ensuring that students are 100% covered financially.