This course will meet in 2 hour blocks three times per week all
advertisement
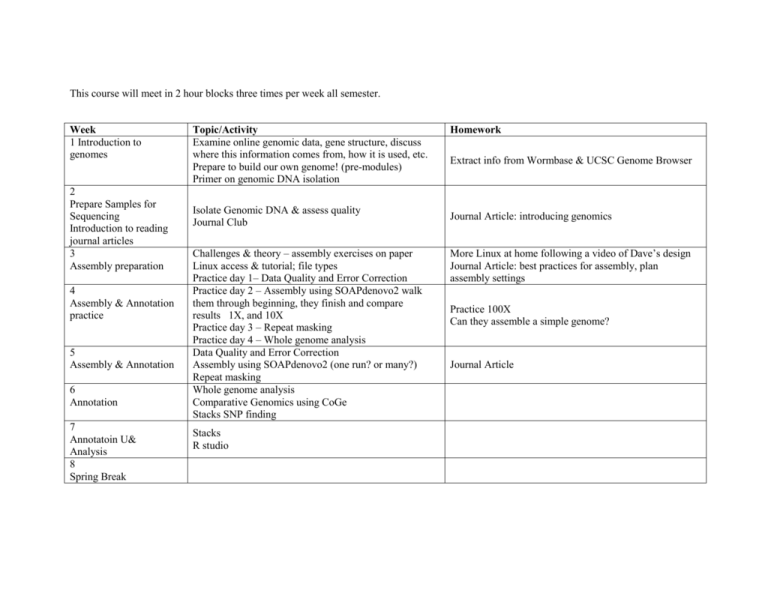
This course will meet in 2 hour blocks three times per week all semester. Week 1 Introduction to genomes 2 Prepare Samples for Sequencing Introduction to reading journal articles 3 Assembly preparation 4 Assembly & Annotation practice 5 Assembly & Annotation 6 Annotation 7 Annotatoin U& Analysis 8 Spring Break Topic/Activity Examine online genomic data, gene structure, discuss where this information comes from, how it is used, etc. Prepare to build our own genome! (pre-modules) Primer on genomic DNA isolation Isolate Genomic DNA & assess quality Journal Club Challenges & theory – assembly exercises on paper Linux access & tutorial; file types Practice day 1– Data Quality and Error Correction Practice day 2 – Assembly using SOAPdenovo2 walk them through beginning, they finish and compare results 1X, and 10X Practice day 3 – Repeat masking Practice day 4 – Whole genome analysis Data Quality and Error Correction Assembly using SOAPdenovo2 (one run? or many?) Repeat masking Whole genome analysis Comparative Genomics using CoGe Stacks SNP finding Stacks R studio Homework Extract info from Wormbase & UCSC Genome Browser Journal Article: introducing genomics More Linux at home following a video of Dave’s design Journal Article: best practices for assembly, plan assembly settings Practice 100X Can they assemble a simple genome? Journal Article 9 Project proposals More detailed annotations? Research Questions & Literature Search Literature workshop 10 Projects Project proposal development Begin projects 11 Projects Generate and analyze project data 12 Projects Interpret and contextualize results 13 14 15 16 Write Manuscript Write Manuscript Draft Manuscripts Work on Presentations Deliver Presentations Final Manuscript annotations ongoing? formulate a good research question peeer review project proposals instructor review project proposals short annotated bibliography work on projects in class and at home write results and begin discussion generate figures tie aspects of manuscript together due at final exam time Access their data set – Technical Challenge: assemble in one run, or each student assemble? Discuss assembly results, N50 score; can compare to Andre’s assembly Phylogeny Exercise ? Annotation – 2 weeks in class, more at home – 50 genes/student in the end? Additional Modules: simple programming in Panther (so they know how some of the software they use works) (Dave can do this ‘easily’)











