Lectures 29 & 30 Notes
advertisement

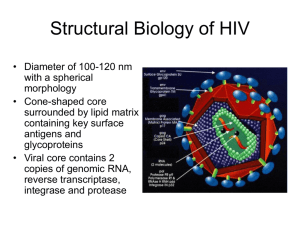
Biology 340 Molecular Biology Lectures 29 and 30 Molecular Biology of HIV May 1 and May 3, 2001 Readings: Trono, D. (1994) Molecular Biology of HIV. Clinics in Laboratory Medicine 14, 203-221. Trono, D. (1996) Molecular Biology and the Development of AIDS therapeutics. AIDS 10 (suppl. 3), 553-559. Outline: 1. Retrovirus life cycle 2. Genome organization of retroviruses 3. Regulation of HIV expression 4. HIV proteins 5. New therapeutic strategies: Brian Watson Lecture: 1. Retrovirus life cycle Retroviruses: Single stranded RNA viruses Covered with lipid envelope Viral genome is copied into DNA then it integrates into host cell genome Three subfamilies; Lentiviruses such as HIV cause slow progressive disorders; Oncovirinae (oncogenic retroviruses) cause many cancers Life cycle: 1. Virus binds to specific cell surface receptor. 2. Virus core is internalized releasing its genome. 3. Viral RNA is copied by reverse transcriptase into double-stranded DNA. 4. The DNA copy of the virus genome integrates into the host DNA. 5. Viral genes are expressed and viral RNAs and proteins are produced and processed. 6. Viral particles are assembled near the cell membrane. 7. New viruses bud from the cell membrane and are released from the host cell. 2. The retroviral genome Two identical chains of RNA Each chain is bound to a tRNA that serves as a primer for reverse transcriptase Ends of genome lack genes, but contain elements (LTR=long terminal repeats) essential for replication, integration and gene expression Encode 3 genes essential for replication gag--structural proteins for virus core pol--genes for viral replication and protein processing: reverse transcriptase, integrase, and protease env--viral envelope glycoprotein HIV-1 contains 6 other genes: vif, vpr, vpu, tat, rev and nef 3. Regulation of HIV gene expression Unique aspects of HIV life cycle: HIV infects cells that express the CD4 receptor: especially helper T lymphocytes and macrophages. Envelope glycoprotein of virus binds receptor. HIV can infect non-dividing cells; most retroviruses infect only dividing cells. This may be related to nuclear import of a viral protein required for assembly. Reverse transcriptase copies of the viral RNA are initiated prior to virus packaging. Genome less than 10 kb, yet expresses nine different genes. Regulation of HIV transcription: Regulation via cis elements in the LTR that bind transcription factors. Viral protein Tat potentiates the effect of cellular transcription factors. Three functional regions of the LTR: modulatory, core, TAR Core region binds basal transcription factors. Modulatory region binds cellular factors such as NF-B. TAR region of viral RNA forms stable stem-loop that Tat binds to. Tat actively recruits cellular transcription factors to the LTR. In presence of Tat, longer transcripts are synthesized--unusual type of regulation. Expression of HIV genes by using overlapping reading frames and alternative splicing. Early genes: tat, rev and nef Late genes: gag, pol, vif, vpr, vpu Rev protein regulates movement of viral transcripts to the cytoplasm for translation. A key determinant of switch from early to late gene expression. Assembly of new viral particles: 1. Viral core made of 1500 gag precursors. 2. Two strands of viral RNA are added. 3. Protease, reverse transcriptase and integrase added to core as a large polyprotein precursor Gag/Pol. 4. Viral envelope glycoprotein is recruited. The glycoprotein is cleaved into two smaller proteins. 5. As virus buds from the cell, the virus protease cleaves the Gag and Gag/Pol precursors into final products. Roles of other HIV-1 proteins: Nef Only found in lentiviruses. Essential for pathogenesis. Regulates cell surface levels of the CD4 receptor. Vif=virion infectivity factor Required for spread of virus Mechanism of action not known Vpr May influence state of differentiation of cells infected by virus Vpu Increases translocation and processing of viral envelope. Enhances release of HIV-1 particles.