1471-2180-12-203-S13
advertisement
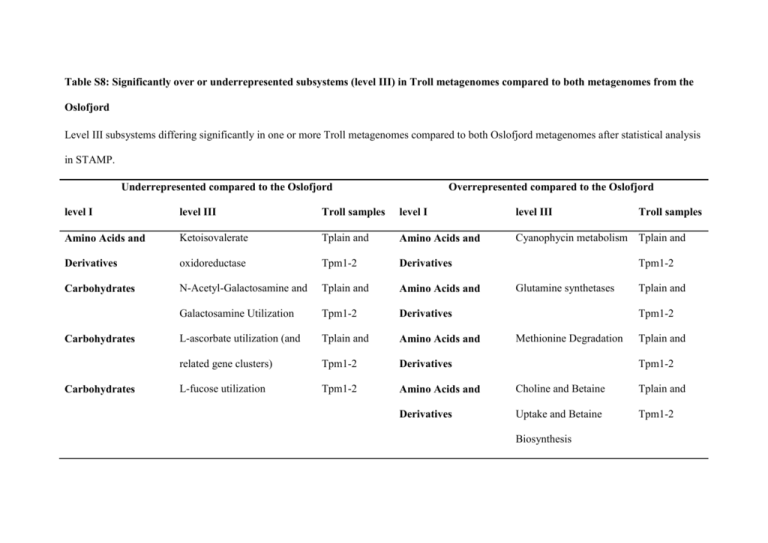
Table S8: Significantly over or underrepresented subsystems (level III) in Troll metagenomes compared to both metagenomes from the Oslofjord Level III subsystems differing significantly in one or more Troll metagenomes compared to both Oslofjord metagenomes after statistical analysis in STAMP. Underrepresented compared to the Oslofjord Overrepresented compared to the Oslofjord level I level III Troll samples level I level III Amino Acids and Ketoisovalerate Tplain and Amino Acids and Cyanophycin metabolism Tplain and Derivatives oxidoreductase Tpm1-2 Derivatives Carbohydrates N-Acetyl-Galactosamine and Tplain and Amino Acids and Galactosamine Utilization Tpm1-2 Derivatives L-ascorbate utilization (and Tplain and Amino Acids and related gene clusters) Tpm1-2 Derivatives L-fucose utilization Tpm1-2 Amino Acids and Choline and Betaine Tplain and Derivatives Uptake and Betaine Tpm1-2 Carbohydrates Carbohydrates Troll samples Tpm1-2 Glutamine synthetases Tplain and Tpm1-2 Methionine Degradation Tplain and Tpm1-2 Biosynthesis Underrepresented compared to the Oslofjord Overrepresented compared to the Oslofjord level I level III Troll samples level I level III Troll samples Clustering-based 1 Tpm1-2 Carbohydrates Methylcitrate cycle Tplain subsystems 1T762T79.3.peg.1T2T62T Clustering-based 1 Tplain Cell Wall and Capsule Lipid A modifications Tplain and subsystems 1T762T80.1T.peg.1T561T Clustering-based 1 subsystems Cofactors, Vitamins, CBSS- CBSS- Tpm1-2 Tplain and Clustering-based 1 2T5731T4.1T.peg.752T Tpm1-2 subsystems 32T0388.3.peg.3759 Flavodoxin Tplain and Clustering-based 1 Tpm1-2 subsystems 2T2T1T988.1T.peg.771T Tpm1-2 Tpm3 Clustering-based GABA and putrescine Tplain and subsystems metabolism from cluters Tpm1-2 Clustering-based 1 Tpm1-2 subsystems 354.1T.peg.2T91T7 CBSS- Prosthetic Groups, CBSS- CBSS- Tplain Tplain and Pigments DNA Metabolism DNA Metabolism ATP-dependent Nuclease DNA repair and recombination eukaryotic Tplain CBSS- Underrepresented compared to the Oslofjord Overrepresented compared to the Oslofjord level I level III Troll samples level I level III Troll samples DNA Metabolism DNA replication, archaeal Tplain Clustering-based 1 Tplain and subsystems 891T87.3.peg.2T957 Tpm1-2 Clustering-based 1 Tplain and subsystems 31T42T69.3.peg.1T840 Tpm1-2 Clustering-based 1 Tplain subsystems 3931T2T4.3.peg.2T657 Tplain and Clustering-based 1 Tpm1-2 subsystems 2T0592T2T.3.peg.1T809 1 and Tpm1-2 Tplain, Tpm1- Cofactors, Vitamins, Chlorophyll Degradation Tpm1-2 1, Tpm1-2, Prosthetic Groups, Tpm2 and Pigments carnitine metabolism Tpm1-2 DNA Metabolism DNA topoisomerases, Type I, Tplain ATP-independent DNA Metabolism Membrane Transport Motility and Late competence ECF class transporters Archaeal Flagellum Chemotaxis Tplain CBSS- CBSS- CBSS- CBSS- Tplain, Tpm1- Tpm3 Nitrogen Metabolism Nitrogen fixation Tpm2 Fatty Acids and Lipids Underrepresented compared to the Oslofjord Overrepresented compared to the Oslofjord level I level III Troll samples level I level III Troll samples Nitrogen Metabolism Nitrosative stress Tpm1-2 Macromolecular 1 Tplain Synthesis 2T432T77.1T.peg.51T1T Metabolism of Biphenyl Degradation Protein Metabolism Glu-tRNA(Gln) Tplain transamidation Protein Metabolism Ribosome LSU eukaryotic Tplain Ribosome SSU eukaryotic Tplain and archaeal Protein Metabolism Translation elongation factors Tplain eukaryotic and archaeal Regulation and Cell signaling Coagulation cascade Metabolism of Phenylpropanoid Tplain and Aromatic Compounds compound degradation Tpm1-2 Metabolism of Benzoate transport and Tplain Aromatic Compounds degradation cluster Metabolism of Gentisare degradation Tplain and Tpm1-2 Aromatic Compounds Tpm1-2 Tplain and Tpm1-2 Aromatic Compounds and archaeal Protein Metabolism CBSS- Metabolism of Salicylate and gentisate Tplain and Aromatic Compounds catabolism Tpm1-2 Underrepresented compared to the Oslofjord Overrepresented compared to the Oslofjord level I level III Troll samples level I level III Troll samples Respiration V-Type ATP synthase Tplain Miscellaneous Biolumenescence Tplain and Tpm1-2 Respiration Respiration Carbon monoxide induced Tplain and hydrogenase Tpm1-2 Reductive Dechlorination Tpm1-2 Nitrogen Metabolism Ammonia assimilation Tplain Nitrogen Metabolism Dissimilatory nitrite Tpm1-2 reductase RNA Metabolism Wyeosine-MimG Tpm1-2 Nitrogen Metabolism Nitric oxide synthase Biosynthesis RNA Metabolism RNA Metabolism RNA polymerase archaeal RNA polymerase archaeal initiation factors Tplain, Tpm12 and Tpm2 Tplain Tplain Regulation and Cell DNA-binding regulatory Tplain and signaling proteins, strays Tpm1-1 Regulation and Cell Orphan regulatory Tplain and signaling proteins Tpm1-2 Underrepresented compared to the Oslofjord Overrepresented compared to the Oslofjord level I level III Troll samples level I level III Troll samples RNA Metabolism RNA polymerase II Tplain, Tpm1- Respiration Terminal cytochrome C Tplain and oxidases Tpm1-2 Biogenesis of Tplain and cytochrome c oxidases Tpm1-2 Protection from Reactive Tplain and Oxygen Species Tpm1-2 Stress Response Flavohaemoglobin Tplain Sulfur Metabolism Inorganic Sulfur Tplain 2 and Tpm3 Stress Response Unclassified Unclassified Rubrerythrin Tpm1-2 Acetone Butanol Ethanol Tplain and Synthesis Tpm1-2 CR clusters Euk 1T Tplain and Respiration Stress Response Tpm1-2 Virulence Adhesion of Campylobacter Tpm1-2 Assimilation Virulence Heme, hemin uptake and utilization systems in GramPositives Tpm2 Unclassified Acyclic terpenes Tplain and utilization Tpm1-2 Underrepresented compared to the Oslofjord Overrepresented compared to the Oslofjord level I level III Troll samples level I level III Troll samples Virulence Pseudaminic Acid Tplain, Tpm1- Unclassified PQQ-dependent Tplain and Biosynthesis 2 and Tpm3 quinoprotein Tpm1-2 dehydrogenases Virulence Bacterial Endolysins: Tpm1-2 Unclassified YgfZ-Fe-S clustering autolysins, phage, and phage- Tplain and Tpm1-2 like lysins Virulence Listeria phi-A1T1T8-like Tpm1-2 prophages Virulence Staphylococcal phi-Mu50B- Hemin transport system Tplain and Tpm1-2 Tpm1-2 like prophages Virulence Arsenic resistance Tplain Virulence Streptolysin S Biosynthesis Tpm1-2 and Transport Virulence 1 Clustering-based subsystems: subsystems in which there is functional coupling evidence that genes belong together, but their exact function is not known.