rrn handout I
advertisement
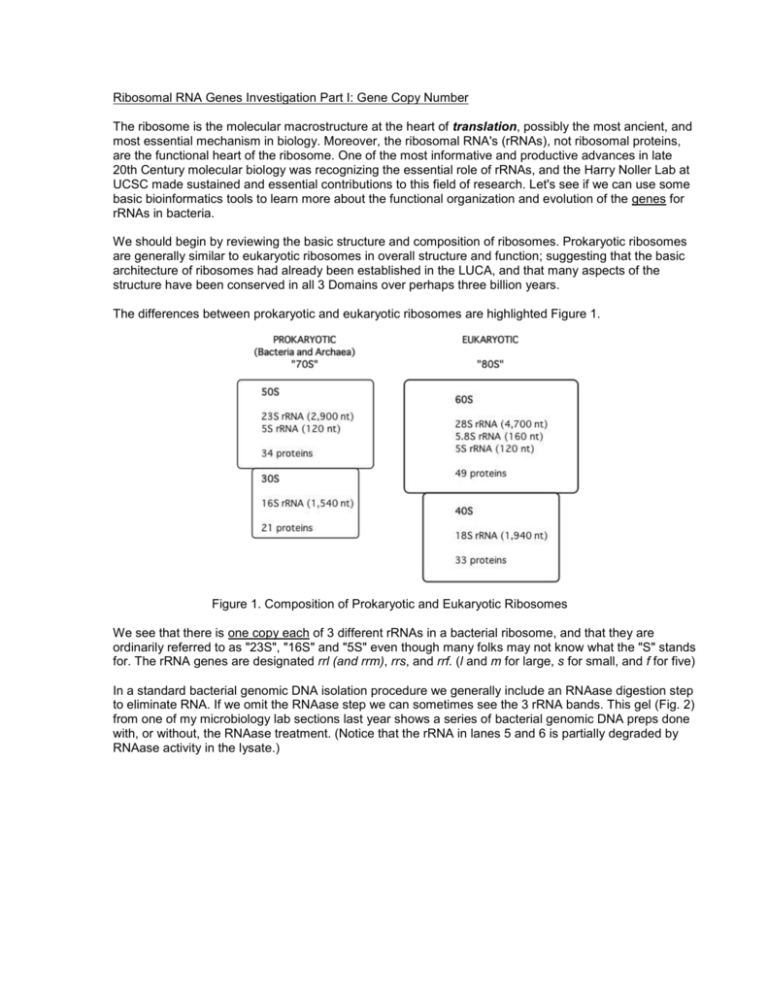
Ribosomal RNA Genes Investigation Part I: Gene Copy Number The ribosome is the molecular macrostructure at the heart of translation, possibly the most ancient, and most essential mechanism in biology. Moreover, the ribosomal RNA's (rRNAs), not ribosomal proteins, are the functional heart of the ribosome. One of the most informative and productive advances in late 20th Century molecular biology was recognizing the essential role of rRNAs, and the Harry Noller Lab at UCSC made sustained and essential contributions to this field of research. Let's see if we can use some basic bioinformatics tools to learn more about the functional organization and evolution of the genes for rRNAs in bacteria. We should begin by reviewing the basic structure and composition of ribosomes. Prokaryotic ribosomes are generally similar to eukaryotic ribosomes in overall structure and function; suggesting that the basic architecture of ribosomes had already been established in the LUCA, and that many aspects of the structure have been conserved in all 3 Domains over perhaps three billion years. The differences between prokaryotic and eukaryotic ribosomes are highlighted Figure 1. Figure 1. Composition of Prokaryotic and Eukaryotic Ribosomes We see that there is one copy each of 3 different rRNAs in a bacterial ribosome, and that they are ordinarily referred to as "23S", "16S" and "5S" even though many folks may not know what the "S" stands for. The rRNA genes are designated rrl (and rrm), rrs, and rrf. (l and m for large, s for small, and f for five) In a standard bacterial genomic DNA isolation procedure we generally include an RNAase digestion step to eliminate RNA. If we omit the RNAase step we can sometimes see the 3 rRNA bands. This gel (Fig. 2) from one of my microbiology lab sections last year shows a series of bacterial genomic DNA preps done with, or without, the RNAase treatment. (Notice that the rRNA in lanes 5 and 6 is partially degraded by RNAase activity in the lysate.) Fig. 2 Agarose Gel Analysis of Genomic DNA preparations form E. coli, with/without RNAase treatment Bacterial rRNA genes are an important exception to the general rule that bacterial genes occur as singlecopy genes. Think about the fact that even though the rRNAs and the ribosomal proteins are needed in equimolar amounts in the cell, the genes coding for ribosomal proteins are present as a single copy, whereas rRNA genes are often present in multiple copies. Fig. 3 shows the variability in copy number for 16S rRNA genes in several hundred prokaryotes. Fig. 3 Distribution of rrs gene copy numbers from the rrn Database (Feb. 2010) It has long been known that the abundance of ribosomes in an E. coli cell is proportional to its growth rate. More recent experimental evidence suggests that rRNA gene copy number is correlated with the ability of prokaryotic cells to achieve rapid growth rates in favorable environments i. i.e. Rapid cell growth requires efficient production of ribosomes, and this is facilitated in turn by having multiple copies of rRNA genes. Lets see if we can find out where E. coli fits in the rRNA gene copy number story. Go to the rrnDB web siteii. You can follow the link from the resources page of the course web site. At the rrnDB Home Page first click on Learn more about rrnDB and then read the brief commentary. Now perform a Search by Taxonomy on FAMILY = "Enterobacteriaceae". This returns a list of all strains, of all species, in all genera, of the Family Enterobacteriaceae for which the database has an entry. (You may need to click "Show All".) This is by no means a complete, representative or unbiased sample of organisms in the Family. It is heavily populated by E. coli strains and by the human pathogens such as Salmonella. It might be helpful to click on the first column heading to sort the entries alphabetically by genus name. You can copy the results and paste them into an Excel spreadsheet for easier manipulation. Explore the dataset by first finding copy #s of the genes for the 23S, 16S and 5S rRNA's in the genome of the standard E. coli K-12 laboratory strain MG1655 (this is the strain we have been designating SC 071). How does this fit in the overall copy number distribution observed for the Domain Bacteria? Spend time looking at, and thinking about, the rRNA gene copy numbers in E. coli and in other members of its Family; paying particularly close attention the variation or distribution pattern of gene copy number. i.e. You should observe that it doesn't look like a table of random numbers! (You can ignore the ITS and tRNA gene copy number information.) Perhaps a frequency histogram would help you visualize this. Try to draw some (tentative) conclusions about the evolution of rRNA gene copy number in the Enterobacteriaceae. Note: All lineages within the Family Enterobacteriaceae diverged from a common ancestor that was present 100 million years ago. WRITTEN ASSIGNMENT Write an organized discussion of what you have found. Begin by summarizing and describing what you have learned from the database. After that proceed to discuss what it may mean in terms of evolution of the bacterial species in the Family Enterobacteriaceae. Don't be tempted to transition too quickly from exploring the database to vague speculation about evolution. Keep you proposals well grounded in the data. Yes, this is somewhat vague and open-ended. That is the point. REFERENCES (Posted on the Web site) i Zarraz May-Ping Lee, Carl Bussema III and Thomas M. Schmidt (2008) rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea Nucleic Acids Research, 2008, 1–5 ii Visualization of ribosomal RNA operon copy number distribution Rajat Rastogi, Martin Wu, Indrani DasGupta and George E Fox (2009) BMC Microbiology 9:208 doi:10.1186/1471-2180-9-208 3 Klappenbach JA, Dunbar JM, Schmidt TM. (2000) rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000 Apr;66(4):1328-33.