Click here to get the file
advertisement

Proposal to create new LOINC codes for supplemental studies in support
of Pathology Biomarker report messages
W. Ted Klein MS; Klein Consulting Inc.
Richard Moldwin MD, PhD; College of American Pathologists
Jaleh Mirza MD, MPH; College of American Pathologists
Created February 21, 2014
Finalized February 24, 2014
Summary:
This proposal requests the code to indicate a report that contains information on supplemental studies on
a cancer case, such as biomarkers. It becomes part of the case report. When carried in the same
message as a primary report (and other supplemental reports, such as Consult reports), it is part of the
Pathology Report Collection labeled panel in the v2 message (60567-5 Comprehensive pathology report
panel). A second proposed code is for a narrative biomarker report, carrying similar information but not
synoptic. Both of these new proposed codes will be additional components of this panel with a cardinality
of 0…n.
Requested code detail - synoptic
Component:
Supplemental Report.synoptic
Long Common name:
Supplemental Report.synoptic
Short Common name:
Supplemental synoptic report
Property:
Finding
Time:
PT
System:
{Setting} (unsure if this should be this for the lab or other place that the source
data came from, or it should be ^Patient, because it is related to studies done
with respect to a Patient.)
Scale:
Doc
Method:
Pathology
Class/Type:
PATH/lab
Member of these panels: 60567-5
Term definition/description:
A supplemental study report is a report that includes supplemental or special studies for a particular
pathology case, such as a biomarkers study. It may be done by a pathologist or laboratory, and serves as
supplemental information to the primary pathology report of the case.
Two additional more granular codes are requested to support biomarker reports.
Both of these are urgently needed by Canada Health Infoway, and are needed asap for use to replace the
‘temporary’ Canadian codes (“XCAnnnn”) in the pCLOCD. It would be very beneficial to get genuine LOINC
codes for these as quickly as possible.
The second code request is:
Requested code detail – synoptic for biomarkers
Temporary code:
XCA03043 (Currently this local code is used in pCLOCD as ‘Biomarker Synoptic
Report’)
Component:
Biomarker Report.synoptic
Long Common name:
Biomarker Report.synoptic
Short Common name:
Biomarker synoptic report
Property:
Time:
System:
Scale:
Method:
Class/Type:
Member of these panels:
Finding
PT
{Specimen} (or should this be ^Patient?)
Doc
PATH/lab
60567-5
The third code requested is for a narrative biomarker pathology report.
Requested code details – narrative for biomarkers
Temporary Code:
XCA03044-5
Component:
Biomarker Report
Long Common name:
Biomarker Report
Short Common name:
Biomarker Report
Property:
Finding
Time:
PT
System:
{Specimen} (or should this be ^Patient?)
Scale:
Doc
Method:
Class/Type:
PATH/lab
Member of these panels: 60567-5
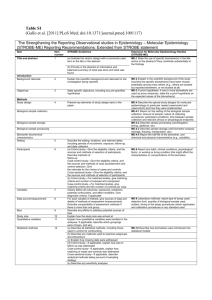
The model for the collection of reports can be seen below:
From NAACCR Volume V version 4.0 Registry Reporting Domain Model
Discussion
The new proposed code will be an additional specialization of the ‘Supplemental Report’ class. The initial
use of this for the CAP checklists is to carry the synoptic biomarkers checklist report. It is intended to be
used in the OBR-4 of a message (where each of the synoptic question/answer pairs are in OBX segments
following). The initial use of this code will be to carry CAP Biomarker checklist information. Note that the
specific measurements from the genetics laboratory for the tests actually run (the LOINC codes for the
many different specific types of EGFR, KRAS, ALK, and other tests) are generally carried as detailed
laboratory results in a separate HL7 ORU message. The registries typically do not wish to receive the
detailed lab results (but they are sent to the pathologists and oncologists treating the patient), but rather
receive the abstracts that are the templates filled in by the Pathologists in crating the synoptic reports.
These supplemental study pathology reports are thus a filtered abstract from the pathologist based on
the information received from the laboratory with the details of the special studies.
Example of the use of the new code
Primary report (60568-3 Synoptic Report)
LUNG: Resection
Synoptic Report Example
Specimen
Specimen: Lung
Procedure: Major airway resection
Specimen Integrity: Intact
Specimen Laterality: Right
Tumor Site: Upper lobe, Middle lobe, and Lower lobe
Tumor Focality: Separate tumor nodules in same lobe
Tumor
Histologic Type: Adenocarcinoma, acinar predominant
Histologic Grade: G2: Moderately differentiated
Extent
Tumor Size
Dimension(s): 3 x 2 x 2 cm
Visceral Pleura Invasion: Not identified, comment for not identified cases
Tumor Extension: Tumor involves main bronchus 2 cm or more distal to the carina,
Parietal pleura, and Chest wall
Involved Chest Wall Structure(s): list of involved chest wall structures
Margins
Bronchial Margin: Not applicable
Vascular Margin: Not applicable
Parenchymal Margin: Not applicable
All Margins Uninvolved by Invasive Carcinoma
Distance of Invasive Carcinoma From Closest Margin: 2 mm
Note: For reporting cancer biomarker testing results, the CAP Lung Biomarker Template should be used.
Pending biomarker studies should be listed in the Comments section of this report.
…
Supplemental Report (new requested code):
LUNG: Biomarker Reporting Template
Example of patient synoptic biomarker report:
Specimen Adequacy
Adequacy of Sample For Testing: Adequate
Estimated % Tumor Cellularity: 2%
Results
EGFR
EGFR Mutational Analysis: No mutation detected (wild-type EGFR allele)
EGFR Exons Assessed: 19
ALK
ALK Rearrangement: No rearrangement detected
Polysomy: Present
KRAS
KRAS Mutational Analysis: No mutation detected
Other Markers Tested
Marker: marker 1
Methods
Sequencing
Whole Genome or Exome Sequencing: Whole genome sequencing - WGS1
EGFR
EGFR Mutational Analysis Testing Method(s): Pyrosequencing - exon 19
ALK
ALK Rearrangement Testing Method(s): In situ hybridization (fluorescence [FISH] or chromogenic
[CISH]): FISH
KRAS
KRAS Mutational Analysis Testing Method(s): Pyrosequencing - codon 12
Testing Method For Other Markers
Other Testing Method: ARMS