Supplemental Digital Content 1. Methods Screening for Point
advertisement

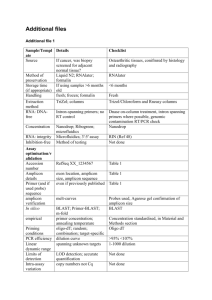
Supplemental Digital Content 1. Methods Screening for Point Mutations and Micro-Insertions/Micro-Deletions in the CASR Gene All six coding exons (i.e., exons 2-7) of the CASR gene were analyzed by direct sequencing for point mutations and micro-insertions/micro-deletions, with the primer sequences and amplicon sizes being supplied below: Exon Amplicon size (bp) Primer Sequence (5 to 3') 2 Forward: GTTGCAGTGGTCATCACAAG 570 Reverse: TATTTTGCGTTTGGTGCAG 3 Forward: CGATGATTCAAACCCAGCT 448 Reverse: CTGCTTCTTCTGATCCTGC 4 Forward: AGAAAGCCACCTCCACAACA 1030 Reverse: GCAGCCCAACTCTGCTTTAT 5 Forward: TGGGGCTTGTACTCATTCTT 290 Reverse: CTGGTTTTCTGATGGACAGC 6 Forward: GCCCAACGTCTGTCACACT 324 Reverse: TCCATGGGCTTCACTGAC 7 Forward: ACACATTTTAGTCTGTGCC 1588 Reverse: CTCCCTAGCCCAGTCTT PCR pertaining to this analysis was performed in a 15 µL reaction mixture using the AmpliTaq® DNA polymerase (Applied Biosystems, Foster City, CA) with 10 ng genomic DNA according to the manufacturer’s protocol. The PCR programme comprised an initial step of denaturation at 94°C for 2min, followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec or 90 sec (according to the amplicon size), and a final extension at 72°C for 10 min. Amplicons were purified by ExoSAP-IT (GE Healthcare, Orsay, France) and sequenced using the ABI PRISM Big-Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). Screening for Gross Genomic Rearrangements or Copy Number Variation in the CASR Gene Gross genomic rearrangements or copy number mutations in the CASR gene were screened for by means of quantitative fluorescent multiplex-PCR. Seven primer pairs were designed to span its seven exons with a control primer pair targeting the MGAM gene located on chromosome 7: Exon Primer sequence (5 to 3') 1 Forward: TGCAGACCTGCCATGCCAATT Reverse: TGTGCGGCCCATATACTTGGA Forward: ACTCAAGGACCACCCACAT Reverse: AATGCCATGGTTCTGCCGT Forward: ATTTCCGTGGGTTTCGCTGGT Reverse: AGGCCTTAGAAACGGTGTTGC Forward: TGGGCACAATTGCAGCTGAT Reverse: ACGATGACTTTGGCCGTGGAAT Forward: AGATGGCTCCATCGTGTT Reverse: AGAACCCACTCCACAGGAT Forward: TCACCTTTGTGCTGTCTGTCCT Reverse: ACACACTCAAAGCAGCAGGT Forward: TCCTGCATTGCCAAGGAGAT 2 3 4 5 6 7 Primer concentration (µM) 0.4 Amplicon size (bp) 154 0.4 209 0.2 132 0.4 178 0.4 104 0.4 115 0.6 142 MGAM exon 7 Reverse: TGACAATGGGTGTGTTGCGGA Forward: TTTGCTGACCAGTTCTTGCAGCTC Reverse: GGGCCCATTTGAAAGCTTACTCCA 0.1 164 All eight targeted regions were amplified in a single PCR reaction with 100 ng genomic DNA in a 10 µL reaction mixture using the Qiagen Multiplex PCR Kit (Qiagen, Courtaboeuf, France). After an initial step of denaturation at 95°C for 15 min, the PCR program comprised 25 cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 60 sec and extension at 72°C for 90 sec, with a final extension at 72°C for 30 min. Amplified DNA fragments were separated on an ABI Prism 310 sequencer (Applied Biosystems, Foster City, CA, USA) and data were analysed with Genemapper v3.4 (Applied Biosystems). Peak heights of the seven CASR amplicons for each patient sample were first normalized against the peak height of the control MGAM amplicon before being superimposed upon those of a control sample. The presence of a deletion was indicated by the two-fold reduction of the corresponding peak. Statistical Analysis Variant frequencies in cases and controls were compared using the χ2 test wherever appropriate. In case of significant differences (P < 0.05), the strength of the association was assessed by calculating the odds-ratio (OR) with 95% confidence interval (CI).