The interactions of bis-phenanthridinium * adenine
advertisement

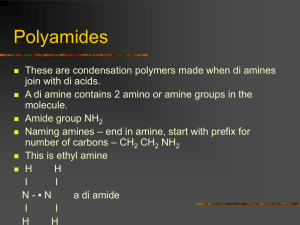
The interactions of bis-phenanthridinium – adenine conjugates with polynucleotides Lidija-Marija Tumir, Filip Šupljika, Ivo Piantanida 1 2 3 Scheme S1. Conformations of studied compounds obtained by MD simulations. (1) Table S1. Groove widths and depths for selected nucleic acid conformations.(2,3) Groove width [Å] Groove depth [Å] major minor major minor a poly dAdT – poly dAdT 11.2 6.3 8.5 7.5 b poly dA – poly dT 11.4 3.3 7.5 7.9 c poly rA- poly rU 3.8 10.9 13.5 2.8 a poly dGdC – poly dGdC 13.5 9.5 10.0 7.2 a B-helical structure (e.g. B-DNA); b C-helical structure (e.g. C-DNA). c A-helical structure (e.g. A-DNA). Table S2. Stability constants (logKs)a for complexes of 3 (4) (citrate buffer, pH = 5.0, c = 0.03 mol dm-3) at pH 5 with ss- and ds-polynucleotides calculated by processing of fluorimetric titrations by means of Scatchard equation (5) 3 poly dA – poly dT 5.4 poly dAdT – poly dAdT 6.2 poly dG – poly dC 6.2 poly dGdC – poly dGdC b poly rA–poly rU 5.3 poly rAH+ – poly rAH+ 5.6 poly U b a Values of log Ks are recalculated according Scatchard equation for fixed n=0.1 for easier comparison; coefficients of correlation were >0.989-0.999 for all calculated Ks; processing of titration data by means of Scatchard equation gave values of ratio n=0.10.05 for most complexes. bThe emission changes at excess of polynucleotide were too small or linear for accurate calculation of binding constants. Thermal melting studies Table S3 Tm-valuesa,b (°C) of different ds-polynucleotides with 1-2 at different ratios rc; pH = 5.0 (I=0.03 mol dm-3, sodium citrate buffer). 1 2 rc = 0.1 0.2 0.3 0.1 0.2 0.3 poly dAdT - poly dAdT 5.7 5.5 5.5 5.4 5.5 5.9 poly A - poly U d -1.2 / 0 d -1.0 / 0 d -1.5 / 0 d 0.9 / 0 d 0.9 / 1.2 d 1.5 / 0 poly dA - poly dT 1.8 2.4 3.5 0.8 1.1 0.8 poly AH+ – poly AH+ 1.1 2.3 4.6 0.6 0.9 1.0 a Error in Tm : 0.5°C; b values were corrected for absorbance of examined compound at different ratios r c r = [compound] / [polynucleotide]; d biphasic transitions: the first transition at Tm = 28.5 oC was attributed to denaturation of polyA-polyU and the second transition at Tm = 80.1 oC was attributed to denaturation of poly AH+-poly AH+ since poly A at pH=5 is mostly protonated and forms ds-polynucleotide. (3) Isothermal titration calorimetry (ITC): Isothermal titration calorimetry (ITC) is used to characterize the thermodynamics of interactions of small molecules with macromolecules (DNA, RNA), providing valuable information about the enthalpic and entropic contributions to the binding Gibbs energy. The data points in figures S1 and S2 reflect the experimental injection heats while the solid lines reflect calculated fits of data. Fitting were performed using the one set of binding site model. 0,5 7 0,0 6 0,0 5 (H / n(1)) / kJ mol -1 -0,5 (H / n(1)) / kJ mol -1 -1 (H / n(1)) / kJ mol -0,5 -1,0 4 3 2 -1,0 -1,5 -1,5 1 0 -2,0 0,0 0,1 0,2 0,3 0,4 -2,0 0,0 0,0 n(1) / n(poly dAdT - poly dAdT) 0,1 0,2 0,3 0,4 0,1 0,2 0,3 0,4 n(1) / n(poly rA - poly rU) n(1) / n(poly dA - poly dT) 8 4 6 (H / n(1)) / kJ mol (H / n(1)) / kJ mol -1 -1 3 4 2 1 0 0 0,0 2 0,1 0,2 0,3 0,4 0,0 n(1) / n(poly dGdC - poly dGdC) 0,1 0,2 0,3 0,4 n(1) / n(poly dG - poly dC) Figure S1. ITC titrations of polynucleotides with the compound 1, (sodium cacodylate/HCl buffer, pH = 5.0, c = 0.05 mol dm-3). Data points are the experimental injection heats while the solid lines represent fitting to one set of binding sites model. The best fits gave values of ratio n[bound compound] / [polynucleotide] = 0.1 0.05 for the most complexes, for easier comparison values of log Ks are recalculated for fixed n = 0.1. 6 0 5 (H / n(2)) / kJ mol (H / n(2)) / kJ mol -1 -1 -1 -2 -3 4 3 2 1 -4 0 -5 0,0 0,1 0,2 0,3 0,4 0,0 n(2) / n(poly dAdT - poly dAdT) 0,1 0,2 0,3 0,4 n(2) / n(poly dA - poly dT) Figure S2. ITC titrations of polynucleotides with the compound 2, (sodium cacodylate/HCl buffer, pH = 5.0, c = 0.05 mol dm-3). Data points are the experimental injection heats while the solid lines represent fitting to one set of binding sites model. The best fits gave values of ratio n[bound compound] / [polynucleotide] = 0.1 0.05 for the most complexes, for easier comparison values of log Ks are recalculated for fixed n = 0.1. The equilibrium constant, binding stoichiometry and reaction enthalpy change were determined directly from the experiment, while entropy contribution and Gibbs energy change were calculated from the calorimetric data. It should be noted that relatively high binding constants with low heat changes didn’t allow accurate analysis of thermodynamic binding parameters, thus data given in Table S4 should be considered as estimations. Table S4. Data parameters observed during fitting with the model one set of sites for ITC titration of polynucleotide with the compound 1 (fixed N = 0.1) log Ka ΔrH/kJ mol-1 ΔrS/J K-1 mol-1 T ΔrS/kJ mol-1 ΔrG/kJ mol-1 p(dAdT)2 5.8 -2.1 103 31 -33 pdApdT 5.6 8.1 134 40 -32 p(dGdC)2 5.3 12.0 141 42 -30 pdGpdC 6.3 3.8 134 40 -36 pApU 5.4 -2.8 93 28 -31 CD experiments 4 4 3 2 2 +poly dG-dC - poly dG-dC 3 1 + poly dGdC - poly dGdC 2 1 1 CD [mdeg] CD [mdeg] 0 -1 -2 poli (dG-dC)2 r = 0.05 r = 0.11 r = 0.21 r = 0.31 r = 0.42 r = 0.52 -3 -4 -5 -6 -7 -8 260 280 300 poly dG-dC - poly dG-dC r = 0.05 r = 0.10 r = 0.20 r = 0.30 r = 0.41 r = 0.51 -2 -3 -4 -5 -6 -9 240 0 -1 320 -7 220 340 240 260 280 300 320 340 / nm / nm Figure S3. CD titration of polynucleotides poly dG-dC - poly dG-dC (c = 2.3 10-5 mol dm-3) with 1 (LEFT) and 2 (RIGHT) at molar ratios r = [compound] / [polynucleotide] (pH = 5.0, buffer sodium cacodylate, I = 0.05 mol dm-3). 2 2 2 + poly dA- poly dT CD [mdeg] CD [mdeg] 1 + poly dA- poly dT 0 poly dA-poly dT r = 0.05 r = 0.1 r = 0.2 r = 0.27 -2 0 poly dA-poly dT r = 0.11 r = 0.18 r = 0.24 -2 -4 -4 240 260 280 300 240 320 260 280 300 320 / nm / nm Figure S4. CD titration of polynucleotides poly dA - poly dT (c = 2.0 10-5 mol dm-3) with 1 (LEFT) and 2 (RIGHT) at various molar ratios r = [compound] / [polynucleotide] (pH = 5.0, buffer sodium cacodylate, I = 0.05 mol dm-3). CD [mdeg] 4 2 poly A-poly U r = 0.09 r = 0.15 r = 0.20 r = 0.31 r = 0.43 r = 0.56 6 CD [mdeg] poly A-poly U r = 0.05 r = 0.16 r = 0.36 r = 0.45 r = 0.56 r = 0.75 6 4 2 0 0 1 + poly A- poly U -2 -2 240 260 280 / nm 300 320 2 + poly A- poly U 240 260 280 / nm 300 320 Figure S5. CD titration of polynucleotides poly A - poly U (c = 1.4 10-5 mol dm-3) with 1 (LEFT) and 2 (RIGHT) at various molar ratios r = [compound] / [polynucleotide] (pH = 5.0, buffer sodium cacodylate, I = 0.05 mol dm-3). poly A r = 0.08 r = 0.13 r = 0.20 r = 0.33 r = 0.43 r = 0.56 4 2 poly A r = 0.1 r = 0.15 r = 0.2 r = 0.3 r = 0.4 r = 0.5 8 6 CD [mdeg] CD [mdeg] 6 4 2 0 0 + 1+ poly AH - poly AH -2 240 260 280 300 + 320 + 2 + poly AH - poly AH -2 -4 340 240 260 280 300 320 / nm / nm Figure S6. CD titration of poly AH+ - poly AH+ (c = 9 10-6 mol dm-3) with 1 (LEFT) and 2 (RIGHT) at various molar ratios r = [compound] / [polynucleotide] (pH = 5.0, buffer sodium cacodylate, I = 0.05 mol dm-3). poly U r = 0.07 r = 0.11 r = 0.22 r = 0.34 r = 0.40 r = 0.51 6 poly U r = 0.05 r = 0.1 r = 0.15 r = 0.17 r = 0.23 r = 0.27 4 2 4 CD [mdeg] CD [mdeg] 6 2 0 0 -2 2 + poly U 1 + poly U -2 -4 240 240 260 280 300 320 340 + 260 280 300 320 / nm / nm Figure S7. CD titration of poly U (c = 3.9 10-5 mol dm-3) with 1 (LEFT) and 2 (RIGHT) at molar ratios r = [1] / [polynucleotide] (pH = 5.0, buffer Na cacodylate, I = 0.05 mol dm-3). 1 Tumir, L.-M.; Grabar, M.; Tomić, S.; Piantanida, I., Tetrahedron. 2010, 66, 2501-2513. 2 W. Saenger, Principles of Nucleic Acid Structure, Springer-Verlag: New York, 1983; p. 226. 3 C. R. Cantor, P. R. Schimmel, Biophysical Chemistry, vol. 3,. WH Freeman and Co., San Francisco, 1980, 1109-1181. 4 Grabar Branilović, M.; Tomić, S.; Tumir, L.-M.; Piantanida, I. Mol. BioSyst. 2013, 9, 2051-2062. 5 a) J. D. McGhee and P. H. von Hippel, J. Mol. Biol., 1974, 86, 469; b) G. Scatchard, Ann. N. Y. Acad. Sci., 1949, 51, 660