docx - Bean Beetles
advertisement

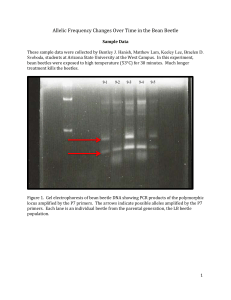
Allelic Frequency Changes Over Time in the Bean Beetle Instructor Notes Experimental Design Any treatment students can think of can potentially skew the allelic frequency, for example population bottleneck, heat treatment, cold treatment, changing bean host types for the beetles, Raid spray. The important thing is to get the students to think about why and how the allelic frequency may change. Bean beetles can be frozen and DNA can be extracted later. Populations (strains) of bean beetles obtained from many sources are homozygous for many if not all genes. Before attempting this experiment, take at least two different strains and cross breed to form a new heterozygous population. Equipment and Supplies For a class of 24, one single experiment could be done to keep costs down. Each PCR will require a single bean beetle to have its DNA extracted, so to do the experiment correctly, a class would need to buy a Qiagen DNAeasy kit with 250 preps. The PCR Platinum Supermix has enough reagent for 100 reactions. To do this experiment correctly, at least 25 bean beetles should be tested before treatment, although more is probably better. Then we tested a few beetles after the treatment, then at least 25 after allowing the population to live for a few generations. If each group wanted to go their own treatment, many kits and PCR Supermix would need to be purchased. Bean Beetle cultures Bean Beetle Bottles Beans Materials for student designed treatment Minipestles Microfuge tubes Qiagen DNeasy Blood and Tissue Kit PBS PCR Primers P7 set Invitrogen PCR Platinum Supermix PCR Machine PCR tubes Gel electrophoresis apparatus Agarose to prepare 1.9% gels TBE or TAE (instructor preference) 1 Ladder DNA for gel reference Bioexpress Gene-Mate Quanti Marker 1kb DNA stain (Gel Red preferred) Phosphate Buffered Saline (PBS) 8 g of NaCl 0.2 g KCl 1.44 g of Na2HPO4 0.25 g of KH2PO4 Bring up to about 900 ml and pH to 7.0. Bring to 1 L, and autoclave. PCR Primers p7 is 5’AGGTTGCAGAAGGAGGCTCT3’ forward and 5’CCGTCCTGGAGCCATATCT3’ backward. This primer pair amplifies a short tandem repeat (STR) that was identified by Smith and Hicks (2012) in their search for polymorphic loci in bean beetles. We bought them from IDT and resuspended them in mpwater at 1g/l. Other PCR primers were evaluated but were not acceptable. The primers mentioned below all amplified but the loci they amplified were not clearly Mendelian in their inheritance. For example, two primers for esterases (Raja et al 2009) amplified and indicated different alleles in two populations but hybrid offspring did not show the expected heterozygous genotype products. RAPD primers described by Fleurat-Lessart and Pronier (2006) (for example P5 in Figure 3 below) amplified but the alleles were not inherited as expected, and other RAPD primers (Gill et al 2006) amplified but were difficult to interpret as alleles in a Hardy-Weinberg equilibrium analysis. 2 Sample Data These sample data were collected by Bentley J. Hanish, Matthew Lam, Keeley Lee, Braelen D. Svoboda, students at Arizona State University at the West Campus. In this experiment, beetles were exposed to high temperature (53°C) for 30 minutes. Much longer treatment kills the beetles. Figure 1. Gel electrophoresis of bean beetle DNA showing PCR products of the polymorphic locus amplified by the P7 primers. The arrows indicate possible alleles amplified by the P7 primers. Each lane is an individual beetle from the parental generation, the LB beetle population. 3 Figure 2. Gel electrophoresis of bean beetle DNA amplified with P7 primers. DNA was extracted from individual beetles and amplified with P7 primers. Lane A1, Africa parental population. Lanes A2-A6, F2 generation of a cross between the LB and Africa population. 4 Lanes 10-14, bean beetle DNA of surviving beetles after 53°C treatment. Arrows indicate three alleles (amplification product sizes). 1000 700 500 200 Figure 3. Representative gel of PCR Analysis with P7 primers of individual beetles 5 generations after the heat treatment. The population seems to have gone to fixation with the 200kb allele (arrow). 5 Data Analysis Usually a chi-squared test is used to determine whether a population is in Hardy-Weinberg equilibrium. Calculating the observed allele and genotype frequencies and expected frequencies with H-W equilibrium are explained in the Student Handout. Using the P7 primer, we analyzed populations of beetles that survived the heat treatment. We saw that the starting P generation either had a genotype of a ~450kb/~350kb, heterozygote or homozygous ~450kb or homozygous ~350kb from the LB population or a single ~200kb allele from the Africa population. After mating the two populations, we saw that the ~200kb was sometimes seen as well in individuals with the heterozygous or homozygous genotype. After treating these beetles with heat, many died, but those that lived, we allowed them to live for about 5 generations (a semester) and then sacrificed them, extracted DNA, and performed PCR with the P7 primers. We saw in all cases that the genotypes of the remaining beetles was only the ~200kb allele. Data Tables Pre-treatment Beetles: 350kb+ 350kb 450kb 18 5 450kb 1 350kb + 200kb 0 450kb + 200kb 0 200kb 8 Post-Treatment Beetles (note students didn’t check very many because they needed the beetles for the rest of the experiment): 350kb+ 350kb 450kb 350kb + 450kb + 200kb 450kb 200kb 200kb 5 0 0 0 0 2 5 Generations Post-Treatment: 350kb/ 350kb 450kb 450kb 0 0 0 350kb + 200kb 0 450kb + 200kb 0 200kb 20 The population seemed to have gone to fixation after the treatment. Population Analysis via Hardy Weinberg Tri-allelic Hardy-Weinberg: p is 350kb q is 450kb r is 200kb 6 Pre-treatment p+q+r=1 p = [18 + 5(2)]/64= 0.44 q = [18 + 1(2)]/64= 0.31 r = [8(2)]/64= 0.25 p2+2pq+q2+2pr+2qr+r2=1 Observed p2=0.16 2pq=0.56 q2=0.03 2pr=0 2qr=0 r2=0.25 sum 18 5 1 0 0 8 32 Expected p2=0.19 0.19(32) = 2pq=0.27 q2=0.10 2pr=0.22 2qr=0.16 r2=0.06 sum Expected Number 6.08 ~ 6 8.64 ~ 9 3.02 ~ 3 7.04 ~ 7 5.12 ~ 5 1.92 ~ 2 32 Chi-squared analysis: (18 − 6)2 (5 − 9)2 (1 − 3)2 (0 − 7)2 (0 − 5)2 (8 − 2)2 + + + + + = Χ2 18 9 3 7 5 2 8+1.78+1.33+7+5+18=41.11 X2=41.11, df=5, p<0.0001, but two genotypes have expected values of less than 5, so the chi-squared test is not appropriate. A Kolmogorov-Smirnov One-Sample test is a better choice when any expected values are less than 5 (www.vassarstats.net). In this case, the observed distribution in the pre-treatment population is significantly different from the expected Hardy-Weinberg equilibrium (n=32, Dmax=0.375, p<0.01). Post-treatment p+q+r=1 0+0+1=1 r=1, the population has been skewed to one allele at this locus. 7 Literature Cited Fleurat-Lessard, F. and Pronier, V. 2006. Genetic differentiation at the inter- and intraspecific level of stored grain insects using a simple molecular approach (RAPD). Proceedings of the 9th International Working Conference on Stored-Product Protection PS5-9 – 6305:446-455. Gill, T.k., Kumri, S., Sharma, V.L., Badran, A.A., Kumari, M. and Sobit, R.C. 2006. Genetic variation in polymorphic males of Callosobruchus maculatus (Coleoptera: Bruchidae) by RAPD-PCR. Cytologia 71(1):57-62. Raja, M., William, S.J., and Hussain, K.J. 2009. Genetic diversity of Callosbruchus maculatus (Fab.) (Coleoptera: Bruchidae) populations. International Journal of Integrative Biology 8(1):15-18. Smith, J. and K. Hicks. 2012. Detecting genetic polymorphisms in different bean beetles populations. www.beanbeetles.org/protocols This experiment was written by Pamela A. Marshall and Lara Ferry (Beanbeetles.org). 8