SUPPLEMENTARY MATERIALS Synthesis of HA-PEI and HA
advertisement

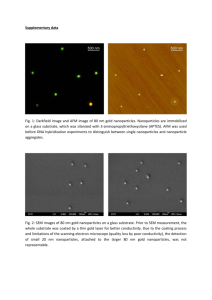
SUPPLEMENTARY MATERIALS Synthesis of HA-PEI and HA-PEG Polymers HA was chemically conjugated with PEI by using 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC). Briefly, HA (MW 20 kDa, 100 mg, 5 mM) was dissolved in 5 mL dry formamide in a glass scintillation vial by incubation at 50°C. The clear solution was allowed to cool to room temperature and 3.3 mg PEI (10 kDa, 0.33 mM) was then added. Subsequently, EDC (10 mg, 50 mM) was added into the solution and the reaction mixture was incubated overnight under magnetic stirring at room temperature. The resulting solution was added to a large excess of EtOH (250 mL) to precipitate the polymer. The precipitate was centrifuged and collected and the washings were discarded. To purify the polymer, EtOH precipitation and washing was repeated thrice. The precipitate polymer was finally re-dissolved in deionized water and subjected to ultrafiltration using Labscale Millipore Tangential Flow Filtration (TFF) system with Biomax 10 kDa MWCO membrane (EMD-Millipore, Billerica, MA). The concentrated and purified polymer was finally lyophilized using a freeze dryer (Freezone-6, Labconco Inc., Kansas, MO) and stored at minus 20 °C (yield: 75 mg, 75%, off-white fibrous product). For the preparation of HA-PEG polymer, HA was chemically modified with poly(ethyleneglycol amine) (PEG2000-NH2) using EDC and n-hydroxy sulfo succinimide (sulfo-NHS). Briefly, EDC (19 mg, 100 mM) and sulfo-NHS (21 mg, 100 mM) were dissolved in 5 mL of deionized water and kept for 1 h to form the amine reactive intermediate. 1 HA (MW 20 kDa, 200 mg, 10 mM) was mixed in 10 ml of deionized water in a glass scintillation vial. After stirring and obtaining a clear solution, 50 mg of the PEG (50 mM, 2 k for 5% modification) was then added to the solution and stirred again. Subsquently, the EDC/NHS solution prepared above was added into the HA/PEG solution drop wise. The reaction mixture was then stirred for 24 h and dialyzed using cellulose dialysis membranes (MW cut off 12 - 14 kDa) against deionized water/methanol mixture (1v/3v - 1v/1v) for another 24 h and finally with deionized water for 48 h. The purified product was lyophilized and stored at minus 20 °C (yield: 80 mg, 79%, off-white fibrous product). 5 mg of the lyophilized product of HA-PEI or HA-PEG was dissolved in 600 μL of D2O and characterized by 400 MHz 1H NMR spectroscopy (Varian Inc., CA) for determining the % modification of the HA backbone. 1H NMR Analysis of HA-PEI and HA-PEG Polymers HA-PEI and HA-PEG derivatives were analyzed for % modification of HA backbone with respective polymers (Fig. S1). The NMR peak at ~1.94 ppm (Blue box; Fig. S1) corresponds to the acetyl groups in the HA backbone, which could be integrated for a total of 141 H (47 monomer residues/HA molecule X 3 H). Similarly, the NMR peak at ~2.7 ppm (Pink box; Fig S1) could be assigned to the two CH2 groups in each PEI monomer and was integrated to 908 H (227 monomer residues/PEI molecule X 4H). The percent substitution for the HA-PEI derivative was thus calculatedand was found to be ~11%. Similarly, the NMR peak at 0.95 ppm (Green box; Fig. S1) could be assigned to the CH3 group in the 2 mPEG molecule. The calculated integration value was assigned to 3 H/mPEG molecule and thus the percent modification was found to be ~7.6% or ~3.7 mPEG molecules/molecule of HA. HA-PEI/HA-PEG/MDR1 siRNA Nanoparticle Formulation and Characteristics The purified of HA-PEI and HA-PEG conjugates (1:1 w/w) were incubated at room temperature for 5 min to form self-assembled nanoparticles (HA-PEI/HA-PEG). We followed a protocol based on our previous research that demonstrated that highest gene silencing efficiency was observed when siRNA was encapsulated in HA-PEI/HA-PEG nanoparticles at a mass ratio of 54:1 (polymer : siRNA). Briefly, MDR1 siRNA was encapsulated in HA-PEI/HA-PEG at a mass ratio of 54:1 (polymer : siRNA) and incubated for another 15 min at room temperature. The particle size and zeta potential measurements of the MDR1 siRNA loaded HA nanoparticles were performed with a Zetasizer Nano S Instrument (Malvern Instruments Ltd, Worcestershire, UK). For light scattering experiments, the samples were measured at a fixed angle of 90o at 25°C. The scattering intensity was adjusted in the range of 50-200 kcps by diluting the samples with 1X PBS. For zeta potentials, default parameters of dielectric constant, refractive index, and viscosity of water were used based on the electrophoretic mobility of the nanoparticles. Transmission electron micrographs (TEM) of nanoparticles were assessed using JEOL JEM-1000 instrument (JEOL Ltd, Tokyo, Japan). 3 Real-time PCR Assay Total RNA was extracted using the Trizlo Reagent® (Life Technologies) according to the manufacturer’s protocol at different time points post transfection. High capacity cDNA reverse transcription kits (Applied Biosystems, Foster City, CA) were used for reverse transcription. Real-time PCR was performed by employing human MDR1TaqMan® gene expression assays (Applied Biosystems) according to the manufacturer’s protocol. Reaction mixtures (20 µL) contained 1.0 μL 20X TaqMan® gene expression assay, 10 μL 2✕ TaqMan® gene expression master mix, 2.0 μL cDNA and 7.0 μL RNase-free water. The PCR cycling conditions were 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 second and then 60°C for 1 min. The real- time PCR was performed using Applied Biosystems StepOnePlus™ System (Applied Biosystems). Western Blot Analysis 20 μg of total protein was processed on NuPAGE® 4–12% Bis-Tris Gel (Life Technologies) and transferred onto the pure nitrocellulose membrane (Bio-Rad). The membranes were then blocked in Odyeesy block buffer for 2 h, and they subsequently incubated with primary antibodies (dilution: 1:1000) at 4°C overnight. They were then further probed with respective secondary antibodies (LI-COR Bioscences, Lincoln, NE) at 1:20000 dilutions for 1 hour at room temperature. The membranes were scanned using Odyssey® CLx equipment (LI-COR Bioscences) after rinsing with TBST three times. 4 Immunofluorescence Assay Briefly, the cells were fixed in 3.7% buffered paraformaldehyde for 15 min and then permeated with methanol for another 20 min. After blocking cells with 1% BSA in PBS for 1 hour, they were incubated with Pgp antibody (dilution: 1:50) overnight at 4°C. On the following day, cells were probed with Alexa Fluor 594 (Red) conjugated goat anti-mouse antibody (Invitrogen) at the dilution of 1:1000 for half an hour. Before taking images, the cells were incubated with 1 μg/mL Hoechst 33342 (Life Technologies) for nuclei staining. Images were acquired by Nikon Eclipse Ti-U fluorescence microscope (Nikon Instruments) equipped with a SPOT RT digital camera (Diagnostic Instruments). Calcein AM Retention Assay 5×104 OVCAR8TR cells were seeded in a 96-well plate in triplicate and transfected with HA-PEI/HA-PEG/MDR1 siRNA nanoparticles. Cells were then incubated with 0.25 μM calcein AM for 15 minutes at 48 h post transfection. After washing cells with cold culture medium three times, calcein AM intracellular accumulation was determined using SPECTRAmax GeminXS® fluorescent spectrophotometer (Molecular Devices) at absorption wavelength 494 nm and emission wavelength 517 nm, and normalized for cellular protein levels as determined by DC Protein Assay. The absorbance values were normalized against the value of untreated cells. 5 Cytotoxicity Assay Cytotoxicity of paclitaxel in OVCAR8TR cells transfected with HA-PEI/HA-PEG/MDR1 siRNA was assessed by the MTT assay. 2×103 cells were seeded in 96-well plates in triplicate and transfected with HA-PEI/HA-PEG MDR1 siRNA nanoparticles. After 48 h post-transfection, the cells were exposed to different concentrations of paclitaxel for 5 days. Subsequently, 20 μL of MTT (Sigma-Aldrich, St. Louis, MO) was added to each well and incubated for 4 h. Dissolving the resulting formazan products with acid-isopropanol, the absorbance was analyzed on a SpectraMax Microplate® Spectrophotometer (Molecular Devices LLC, Sunnyvale, CA) at a wavelength of 490 nm. Absorbance values were normalized assigning the value of the cells in medium without drug to 1.0 and the value of the blank well (without cells) control to 0. Supplementary Figure legends Fig. S1 1H NMR spectra of HA-PEI and HA-PEG derivatives dissolved in D2O. The blue box indicates the acetyl group on HA backbone while the pink and green boxes indicate the ethylene groups in PEI and methyl group in mPEG respectively that were used for calculation of percent modification. Fig. S2 The cytotoxicity of different polymer vectors in OVCAR8TR cells. HA-PEG polymer 6 was significantly less cytotoxicity than HA-PEI, and the cytotoxicity decreased when HA-PEI (1:1 ratio) was mixed with HA-PEG. Most importantly, the cytotoxicity of HA-PEI/HA-PEG slightly decreased once the MDR1 siRNA was encapsulated in the nanosystem. The experiments were performed in triplicate. Fig. S3 Schematic showing the steps involved in the synthesis of HA-PEI and HA-PEG derivatives and the formation of self-assembled nanoparticles in the presence of MDR1 siRNA. 7