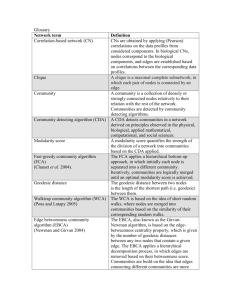
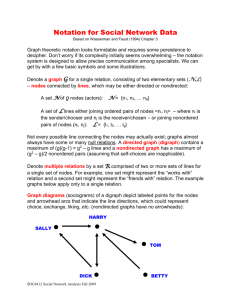
Answers to some network questions from the C+
advertisement

How in nature do the intrinsic vulnerabilities to attack in small world networks manifest in nature? How, if at all, are they controlled? (are there any techniques suggested by small world networks to guard against targeted attacks) This was more a weakness on the part of the presentation – small world networks are actually very robust to random pertubations (think of the airport networks – if an airport is removed at random, the effect is likely to be small). People have hypothesized that one of the reasons small world networks are so common in nature is that they are so very robust to random pertubations. For instance, it might be advantageous for biological systems to be resistant to random changes or infections. Of course, there are therefore also viruses that have evolved to interfere with certain central proteins, as you have mentioned. I’m not sure how these intrinsic weaknesses to random external attacks are controlled; however, from an evolutionary standpoint, researchers have found that the genes which change the most slowly are indeed the ones that regulate many genes. How could small world networks be applied to biomolecular pathways? Small world networks are generally applied in the context of networks of dynamical systems, meaning systems made up of many discrete elements, with each element behaving in accordance with particular rules based on the behavior of the elements around it. With this in mind, it might not be appropriate to model biomolecular pathways as networks. (paraphrased) Is there a different optimum value of the probability of rewiring for all small-world networks? I’m not entirely sure what you mean, since there isn’t an optimum probability in general for small-world networks – in our presentation we showed that there was a particular range of values for which you get interesting and new behavior in networks of dynamical systems. Small world networks are the class of network that you get when the probability of rewiring, p, is tuned to a specific range of values. How is randomness determined in networks? Random in the sense that we meant it here was that each element in the network is equally likely to be connected to every other element – that is, it is not possible to predict from the physical location of a node which nodes it will then be connected to. Mathematically, you take a set of n vertices, and add edges between them at random. Based on the different models for generating these edges, different properties can arise at different stages of adding these random edges. In this case, the process was rewiring a regular network with probability between 0 and 1, which made, as we have seen, a set of graphs with rather interesting properties. How can we apply dynamical networks to study biological systems/can we study macrobiological things with small-world networks/ what kind of things can the small-world network model/what is the biophysical significance of the small-world network? With regard to dynamical networks: most biological systems are just that – systems. In other words, there are elements, which push and pull on each other to varying degrees, which then change the behavior of the components. Making computational models of the way in which these elements push and pull on each other, which then gives rise to interesting sets of behavior, is the first step towards an understanding of these larger complex systems. Small-world networks make a statement about the connectivity of the system (making conclusions based on the examination of a single property of the network, its topology). It makes sense, then, that the things the small world network can model are based mostly on connectivity. The surprise is that after measuring some real world systems, it turns out that many things are connected in this very special way! As we’ve heard in our class lectures, computational models can often be very useful for giving insights to the way in which things work which may not be immediately accessible experimentally. For instance, in order to understand the formation of memory, scientists have built computational models of the neuron system based on small-world networks, and have found that they exhibit properties such as short-term memory. We might then ask the question of whether you can extrapolate properties discovered via computational models to other small-world networks. Theoretical study of these small world networks therefore could point towards interesting new experiments or missing experimental data that might not otherwise be considered. Are there more mathematical models and equations that govern specific networks? If so, are there methods of optimizing these networks to meet specific goals? The small world network is rigorously defined mathematically – in fact, network theory is a well defined branch of mathematics! Like I said in an earlier answer, the small-world network is what happens when you tune one particular parameter – the connectivity. There are other parameters that you could also tune to find different properties. For instance, you could look at the rules that link nodes in a network to each other. There are also many other types of network; for instance the hierarchical network – which some have used to model genetic regulatory networks – which have lower nodes, which are influenced by mid level nodes, which are then influenced by high level nodes. The structure of this particular type of network is well optimized to meet specific goals. For instance, in a genetic regulatory network, you can look at the distribution of nodes across different levels of influence – it turns out that there are relatively few high level nodes which control more mid-level nodes, which then control many low-level nodes. In other words, the nodes with a high level of connectivity are the nodes which control many other nodes. Because evolution operates on the basis of random mutation, this is good – a random mutation that happens to one of the many low level genes will generate variation that isn’t likely to destroy the entire organism. (this is my rather inaccurate recounting of the Physics Colloquium which took place on Wednesday 30th). So if we removed a major city we could stop the spread of disease? That is certainly one conclusion you could draw from the research, given that the rate of spread of disease is closely linked to the geometry of the network….another, of course, would be to concentrate on prevention in the major cities.