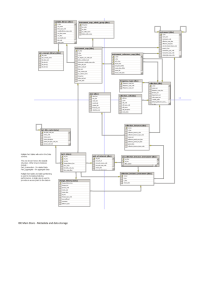
iCORDI-D2 4-0_v4-with changes
advertisement

Project Acronym RDA Europe Project Title Research Data Alliance Europe Project Number 312424 Deliverable Title First year report on RDA Europe analysis programme Deliverable No. D2.4 Delivery Date Author Herman Stehouwer Diana Hendrickx ABSTRACT All detailed analysis results of the data architectures and organizations of the communities studied and a description of possible generalizations towards solutions for integration and interoperability to foster the RDA Europe Forum and RDA discussions. RDA Europe (312424) is a Research Infrastructures Coordination and Support Action (CSA) co-funded by the European Commission under the Capacities Programme, Framework Programme Seven (FP7). 2 rdaeurope@rd-alliance.org | europe.rd-alliance.org 2 3 DOCUMENT INFORMATION PROJECT Project Acronym RDA Europe Project Title Research Data Alliance Europe Project Start 1st September 2012 Project Duration 24 months Funding FP7-INFRASTRUCTURES-2012-1 Grant Agreement No. 312424 DOCUMENT Deliverable No. D2.4 Deliverable Title First year report on RDA Europe analysis programme Contractual Delivery Date 9 2013 Actual Delivery Date 10 2013 Author(s) Herman Stehouwer, MPI-PL; Diana Hendrickx, UM Editor(s) <Insert deliverable editor(s) – Name, Surname, Org Short Name> Reviewer(s) Natalia Manola & Guiseppe Fiameni Contributor(s) Constantino Thanos Work Package No. & Title WP2 Access and Interoperability Platform Work Package Leader Peter Wittenburg – MPI-PL Work Participants CSC, Cineca, MPG, EPCC, CNRS, STFC, UM, ACU, ATHENA, CNR Package Estimated Person Months 16 Distribution public Nature Report Version / Revision 1.0 Draft / Final Draft Total No. (including cover) Pages 47 Keywords rdaeurope@rd-alliance.org | europe.rd-alliance.org 3 4 DISCLAIMER RDA Europe (312424) is a Research Infrastructures Coordination and Support Action (CSA) cofunded by the European Commission under the Capacities Programme, Framework Programme Seven (FP7). This document contains information on RDA Europe (Research Data Alliance Europe) core activities, findings and outcomes and it may also contain contributions from distinguished experts who contribute as RDA Europe Forum members. Any reference to content in this document should clearly indicate the authors, source, organization and publication date. The document has been produced with the funding of the European Commission. The content of this publication is the sole responsibility of the RDA Europe Consortium and its experts, and it cannot be considered to reflect the views of the European Commission. The authors of this document have taken any available measure in order for its content to be accurate, consistent and lawful. However, neither the project consortium as a whole nor the individual partners that implicitly or explicitly participated the creation and publication of this document hold any sort of responsibility that might occur as a result of using its content. The European Union (EU) was established in accordance with the Treaty on the European Union (Maastricht). There are currently 27 member states of the European Union. It is based on the European Communities and the member states’ cooperation in the fields of Common Foreign and Security Policy and Justice and Home Affairs. The five main institutions of the European Union are the European Parliament, the Council of Ministers, the European Commission, the Court of Justice, and the Court of Auditors (http://europa.eu.int/). Copyright © The RDAEurope Consortium 2012. See https://europe.rd-alliance.org/Content/About.aspx?Cat=0!0!1 for details on the copyright holders. For more information on the project, its partners and contributors please see https://europe.rd-alliance.org/. You are permitted to copy and distribute verbatim copies of this document containing this copyright notice, but modifying this document is not allowed. You are permitted to copy this document in whole or in part into other documents if you attach the following reference to the copied elements: “Copyright © The RDA Europe Consortium 2012.” The information contained in this document represents the views of the RDA Europe Consortium as of the date they are published. The RDA Europe Consortium does not guarantee that any information contained herein is error-free, or up to date. THE RDA Europe CONSORTIUM MAKES NO WARRANTIES, EXPRESS, IMPLIED, OR STATUTORY, BY PUBLISHING THIS DOCUMENT. rdaeurope@rd-alliance.org | europe.rd-alliance.org 4 5 GLOSSARY ABBREVIATION DEFINITION RDA Europe Research Data Alliance Europe OAI-PMH Open Archives Initiative Protocol for Metadata Harvesting CSC Finnish IT Centre for Science UM Maastricht University MPI-PL Max Planck Institute for Psycholinguistics CLST Centre for Language and Speech Technology RU Radboud University CNRS Centre national de la recherche scientifique ENVRI Common Operations of Environmental Research infrastructures TNO Dutch Organisation for Applied Research E-IRG E-Infrastructure Reflection Group EEF European E-Infrastructure Forum ESFRI European Strategy Forum on Research Infrastructures ACU Association of Commonwealth Universities CERN European Organization for Nuclear Research MPG Max Planck Gesellshaft rdaeurope@rd-alliance.org | europe.rd-alliance.org 5 6 TABLE OF CONTENTS Executive Summary ....................................................................................................... 7 1 Introduction ............................................................................................................... 8 2 RDA/Europe Forum ..................................................................................................... 8 3 Trends and Gaps ........................................................................................................ 9 #1 Usage of data management .................................................................................... 9 #2 Data models ....................................................................................................... 10 #3 Data preservation anticipation ............................................................................... 10 #4 Availability and quality of available data / metadata ................................................. 10 #5 Metadata ............................................................................................................ 11 #6 discoverability of data .......................................................................................... 11 #7 Data reuse is common, but dependent on field ........................................................ 11 Annex 1: Interview Reports RDA/Europe ........................................................................ 13 Community Data Analysis: EMBL - EBI – molecular databases ........................................ 13 Community Data Analysis: Genedata .......................................................................... 14 Community Data Analysis: TNO .................................................................................. 16 Community Data Analysis: ENVRI ............................................................................... 17 Community Data Analysis: Svali ................................................................................. 24 Community Data Analysis: EISCAT 3D ......................................................................... 27 Community Data Analysis: Public Sector Information ENGAGE ........................................ 30 Community Data Analysis: ESPAS ............................................................................... 33 Workshop: On-line databases : from L-functions to combinatorics .................................. 34 Community Data Analysis: Huma-Num ........................................................................ 37 Community Data Analysis: INAF centre for Astronomical Archives ................................... 38 Community Data Analysis: ML-group........................................................................... 40 Community Data Analysis: Computational Linguistics Group .......................................... 41 Community Data Analysis: CLST ................................................................................. 43 Community Data Analysis: Donders Institute ............................................................... 44 Community Data Analysis: Meertens Institute .............................................................. 45 Community Data Analysis: Microbiology ...................................................................... 47 Community Data Analysis: Arctic Data ........................................................................ 48 rdaeurope@rd-alliance.org | europe.rd-alliance.org 6 7 Executive Summary In this report we show the results for the first year from WP2.3: the analysis program. One of its main components is the presence in the annex of all the interview reports produced under this program. The following organizations and communities have been interviewed and reported: Institutes/companies: TNO (general semi-commercial research institute, toxicogenomics case) Genedata (genetic data) Donders institute (brain research) Meertens Institute (dialects and linguistic history in the Netherlands) Arctic Data Institute (NIOZ) INAF centre for Astronomical Archives EMBL-EBI (Molecular databases) Research Groups/Departments: Machine Learning research group Computational Linguistics research group Language and speech technology group Microbiology research group Other: Math Community (on L-functions and combinatorics) ENVRI (large environmental community) Svali (Artic Lands and Ice) EISCAT 3D (3D Imaging radar for atmospheric and geospace research) ENGAGE (Public Sector Information) ESPAS (near earth space research community) Huma-Num (Very Large Research Infrastructure on numerical science for SSH) Interviewing will continue in the second year of the project and the report will be updated to reflect new insights and to include all interview reports. The analysis of the interview reports shows that we still have a long way to go before good data stewardship is commonplace. Furthermore, it underlines the importance of having good metadata. Good metadata enables discoverability and reuse of the data. Based on the analysis in the report we make a number of concrete suggestions, they are listed in short for below: Use basic data management as outlined in the e-IRG whitepaper Consider data management before data collection takes place Document your data with high-quality metadata Use persistent identifiers Ensure discoverability of the data Share your data Researchers should be more familiar with the available possibilities of cloud computing, grid computing, HPC We recommend the usage of computer-actionable data management policy rdaeurope@rd-alliance.org | europe.rd-alliance.org 7 8 1 Introduction In this report we give an overview of the first year of the RDA Europe Analysis programme in which we have conducted many interviews in order to obtain a better and broader understanding of current data practises. We focus specifically on finding gaps and good existing solutions. This report will be updated as soon as the programme is completed during the second year. The report contains a fair amount of additional material that is relevant. In Annex 1 we provide all the analysis reports that are finished at the time of writing. This report will be updated as new interviews come in. In this report we will first describe the RDA/Europe Forum, as the forum is part of the target audience of this report. We will then describe trends and gaps that we note over multiple reports and our report ends with some concrete proposals to improve data exchange, to foster the RDA Europe Forum and to foster RDA discussions. 2 RDA/Europe Forum The process to establish the RDA/Europe Forum was started by the need to be engaged at the European level. John Wood (ACU) discussed with Kostas Glinos (DGConnect) and Carlos Morais-Pires (DGConnect) the possible membership and the types of key stakeholders. When the iCORDI proposal was written it was not exactly clear what was meant by the RDA/Europe Forum (HLSF at the time), different views existed on this. After the granting of the project it was obvious that a group of people representing organizations relevant to the European scenario are needed, furthermore, a group of people that can give backing to the needs of the project politically. The Commission has had the e-IRG, EEF (http://www.einfrastructure-forum.eu/) and different ESFRI groups but none has been really good in bringing together the e-Infrastructure service providers and researchers. The RDA Europe Forum and the RDA Science workshops are also aligned to this purpose.. We have established the RDA/Europe Forum by inviting organisations to send a representative. The current members of the forum (and the organisations they represent) are: P Ayris (LIBER) Jens Vigen (Eiroforum) Norman Wiseman (Knowledge Exchange) Peter Linton (EIF) Donatella Castelli (ERF) Martin Vingron (MPG) Marja Makarow (ESF) Norbert Lossau (LERU) Sandra Collins (European Academies) Paul Boyle (Science Europe) The RDA/Europe Forum is organized by John Wood (ACU), Leif Laaksonen (CSC), Peter Wittenburg (MPI-PL), and Herman Stehouwer (MPI-PL). rdaeurope@rd-alliance.org | europe.rd-alliance.org 8 9 The forum has had one meeting in the first year of the RDA Europe project on 7 March 2013. The second meeting will take place towards the end of November 2013. We expect to continue with around 2 meetings per year. At the first HLSF meeting the RDA/Europe project, the RDA, the (then) current state of RDA, and the RDA/E Forum itself where discussed. Several recommendations were made by the forum. These recommendations include the following: “Organizations should be able to endorse the RDA work”. I.e. the RDA work is performed by individuals in working and interest groups. The outcomes from these group must be adopted. All groups are required to have some form of adoption strategy, however the forum felt it was useful if there was a way for organisations to state their general support for RDA outputs. “Setting up an international legal entity will be a lot of work”. I.e. several members of the forum have experience in setting up international legal entities. Their experience is that setting up such an entity is a lot of work and takes around a year to set up. In the upcoming RDA/E Forum meeting, we will discuss the following items: The RDA legal entity How to strengthen the utilization of RDA output in Europe Liaising more broadly in Europe Industrial connections The upcoming Plenary meeting (March 2014, Dublin) The upcoming RDA/Europe Science workshop The upcoming G8+O6 meeting; Briefing and feedback. This deliverable will be presented to the forum to provide another overview of the status quo and to allow them to push the concrete proposals made. 3 Trends and Gaps Here we will report a picture of the current practises, technologies, models and standards adopted by the interviewed data organizations. Such a picture is of interest to the RDA activities and can help the working groups. The goal of the interviews was to establish such a picture, to find gaps, and to find existing solutions that could be generalized. These findings are one input to the RDA/Europe forum, can also inform new RDA working groups, and are input to the RDA process in general. #1 Usage of data management Data management is the overall practise of dealing well with data. E.g. defining the lifecycle of pieces of data, defining how data can be found inside of the organization. A clear trend that we observe from these interviews is that the quality of the data management is highly variable. Obviously the data infrastructures practise good data management, but once you drop down to institutional or research group level the usage of data management varies wildly. There is also a need for archives to store the growing amounts of data. In some fields, data are in the order of petabytes and larger. rdaeurope@rd-alliance.org | europe.rd-alliance.org 9 10 The underlying data management systems used differ. Simple relational databases are popular in a large number of the interviewed organizations to store their data. The data in the database are usually linked to the original data in the file system. No clear trend as to data management systems is present. In most cases the system is either human-actions or custom build (e.g. a collection of scripts). The use of computer-actionable policy is rare. Based on this point we recommend the encouragement of basic data management as outlined in the e-IRG whitepaper. To be precise: use standard data formats where possible, if there is no clear need for a new definition; state clearly in the metadata which data format is used; have metadata. #2 Data models With relation to data models we can say that the data models used depend on the field interviewed. Obviously this is not surprising. For measurements and analysis there is a clear practise to store these in some XML format. Often data is stored as a time series, either in the raw data or in a relational database, e.g. the NIOZ stores large time series of the DTP (depth, temperature, pressure; often much richer than just dtp) measurements. The use of simpler models (such as array-based) or unusual models (such a graph-based) is limited, to give an example, the biomedical community in EMBL-EBI uses several array-based data models. #3 Data preservation anticipation Several interviewees mentioned that data storage, exchange and preservation must be considered in a preparatory phase and not as an afterthought. That is, when determining what to measure and store one also has to consider what to do with the data. When producing data, long term aspects have to be taken into account. Data preservation is at the moment problematic, but it is clear that, going forward, communities think of preservation wrt their future requirements. Based on this point we recommend considering data management at an early stage, before any data collection takes place. #4 Availability and quality of available data / metadata For several fields, there is a lack of availability of open data. Several interviewees mention that if metadata is available for data that its quality is usually not good enough to find valuable data based on its metadata. There is also a lack of persistent identifiers for data / metadata and the (meta)data are in different formats. There is a need for standardization. rdaeurope@rd-alliance.org | europe.rd-alliance.org 10 11 Based this point we can recommend that data have to be better documented with high quality metadata. Persistent identifiers have to be used. #5 Metadata Most interviewed organizations used a clear, domain-specific, xml-based, metadata standard, though almost all used different ones. However, metadata is not always produced for all data. It is however clear that OAI-PMH is the protocol that is used for metadata exchange. Many of the organizations interviewed underlined the importance of metadata for discovery. #6 discoverability of data Several participants on the institutional level (e.g. Meertens, NIOZ) mentioned that the discoverability of their data is essential to encouraging external reuse. Larger participants, i.e. the research infrastructures, also take care that the data are discoverable. There is a need for integrated data discovery, but this is hampered by the lack of interoperability between different data sources. Based on this point data should be discoverable and interoperable, first through quality metadata, and second through the use of data standards. Persistent identifiers have to be used. #7 Data reuse is common, but dependent on field Data reuse is inherent to the approach taken by the larger data infrastructures that were interviewed. Furthermore, smaller groups (e.g. CL, CLST, Meertens) all indicated that for them data reuse was extremely common. However the small groups often do not have the infrastructure to advertise the available data so the reuse is often contained within the institute itself. Once the infrastructure is available data reuse is also common with third parties. A caveat here is that in some other fields than those interviewed here, e.g. linguistics, data sharing and reuse is less common. Furthermore, in some cases the data contain sensitive information and their sharing is not possible, for instance when talking about patient data or other medical measurements on subjects. Based on this point we recommend the encouragement of data sharing in all fields, unless data are too sensitive, i.e., privacy issues are involved. Concretely we recommend that metadata is always available, so that it is at least clear which data exists. #8 Use of cloud / grid computing rdaeurope@rd-alliance.org | europe.rd-alliance.org 11 12 There is no general trend of using cloud computing for storing and analysing large amounts of data. Based on this point we recommend that researchers become more familiar with the available opportunities for cloud computing to process the growing amounts of data. Furthermore we can recommend to express data management policy as computer actionable rules, which reduces the possibility of human error, saves a lot of effort and enforces consistency with the rules. rdaeurope@rd-alliance.org | europe.rd-alliance.org 12 13 Annex 1: Interview Reports RDA/Europe Community Data Analysis: EMBL - EBI – molecular databases Goals of analysis By interviewing EMBL – EBI, we hope to learn more about data storage, data management and data warehouses. Analysis provided by Maastricht University (UM). Description of the community EMBL-EBI provides freely available data from life science experiments, performs basic research in computational biology and offers an extensive user training programme, supporting researchers in academia and industry. Goals of the community with respect to data provide freely available data and bioinformatics services coordinate biological data provision throughout Europe Data types EMBL – EBI provides access to data from life science experiments. Submitted data files are mostly in TAB-delimited text format, but some are in XML. For ArrayExpress, (transcriptomics) data are submitted by the user in MAGE-TAB format, or via MIAMExpress submission tool. The following file formats are used: - .gpr, .txt, CEL plus EXP (raw data) - .txt, CHP (normalized data) - .txt (combined data file) Metadata are submitted in tab-delimited files when using MAGE-TAB, and via a web interface when using MIAMExpress. Data flow Data is collected and processed by external institutes. The data is submitted into one of the EMBL-EBI databases. A team of curators receive the submissions and may curate the files manually. Curated data can be used by other research institutes or companies. Data organization EMBL-EBI adopts Data Management Systems: relational database (ORACLE / MySQL). Data are stored in Data Warehouses. Examples are ArrayExpress (transcriptomics: microarrays, RNA-seq), PRIDE (proteomics: Mass-Spec), ENA- SRA (genomics: sequencing, next generation sequencing data), Metabolights (metabolomics: NMR, MS) and project-specific data warehouses such as the metagenomics portal and diXa (toxicology). Data exchange The computer environment of EMBL-EBI is a centralized system. Multiple computers are communicating through a network. EMBL-EBI mirrors data from other databases (e.g., ArrayExpress manages GEO (Gene Expression Omnibus) data – a similar database at NCBI). The data are transformed to the format used by EMBL-EBI and loaded in EMBL-EBI databases. rdaeurope@rd-alliance.org | europe.rd-alliance.org 13 14 EMBL-EBI has a Biological Sample database (BioSamples) for integration of the biological sample dimension of different –omics data (transcriptomics, metabolomics, proteomics). Data services and Tools EMBL-EBI provides several databases for experimental data from the life sciences. A recent EBI website redesign resulted in more consistent interfaces of different EBI databases; this process is still ongoing. If the user wants to submit data into one or more of the EMBL – EBI databases, he has to register by creating an account for each database. Further EMBL-EBI provides tools to support the development and use of ontologies. The Experimental Factor Ontology (EFO) provides a systematic description of many experimental variables available in EBI databases, and for external projects. It combines parts of several biological ontologies. The scope of EFO is to support the annotation, analysis and visualization of data. Legal and Policy Issues All data are open. If the user submits data, there is an option to keep data private for a certain period before it becomes public. This option is normally used when data accompany a publication that is under review, in which case manuscript reviewers are able to access the dataset before it becomes public. There are no restrictions to data re-use. An exception is the European Genome-phenome Archive (EGA) which serves as a broker of patient data where data access is carefully managed. Limitations /issues EMBL-EBI doesn’t adopt a generic service for Persistent Data Identifiers, however, it guarantees the persistence of data stored in production repositories. EMBL-EBI currently does not have a database that connects data in different databases for multi-omics studies. Proposed actions EMBL-EBI through the diXa project is currently talking to EU-infrastructures that provide services for persistent identifiers (EUDAT). EMBL-EBI is working on a BioStudy database that connects data in different databases for multi-omics studies. Community Data Analysis: Genedata Goals of analysis By interviewing Genedata, we hope to learn more about data services and data integration. Analysis provided by Maastricht University (UM). Description of the community Genedata is a bioinformatics company that provides scalable enterprise software solutions and consultancy. Goals of the community providing consultancy for data analysis in the life sciences; developing software platform for analyzing and visualizing large experimental data sets generated from life science experiments; rdaeurope@rd-alliance.org | europe.rd-alliance.org 14 15 Data types Genedata develops software platforms for the management and analysis of data from life science experiments. The data are from (among others) –omics platforms, clinical labs and production environments. Input data formats are vendor (platform) dependent; parsers are available for all major platforms. Meta-data formats are either tab-delimited, or standard formats (e.g. MAGE-ML, ISA-Tab). Data are stored internally in relational databases (Oracle), or binary files. Data export can be in done in tab-delimited files (e.g. GGF genomic feature format), Excel files, binary files, or pdf reports. Data flow Genedata supports different stages of the data life cycle: experimental design, data preprocessing, quality control, data management, data analysis, data visualization, and result interpretation. For data preprocessing, quality control and data analysis, the client can use one of the software modules developed by Genedata or consult Genedata for support. Data organization Genedata seamlessly integrates into in-house Data Management Systems (e.g. LIMS) as well as custom relational database (ORACLE/SQL). Open file formats / open interfaces are used as much as possible. Own formats are published. All formats are documented. Data exchange The software system is based on a client server architecture. The programming language for the client and server is JAVA and therefore mostly platform independent. The architecture is a classical three tier architecture: application server, database server (ORACLE), windows client (WebStart). The vast majority of the data stay on the server. Genedata adopts a symmetric multiprocessing (SMP) architecture, Grid technologies (SUN grid engine/Open Grid Scheduler/DRMAA). Data services and Tools Genedata uses Lightweight Directory Access Protocol (LDAP) and Active Directory (AD) for authentication / authorization. Genedata provides a huge list of algorithms / methods for data discovery, data analysis, data mining and data visualization. Genedata software allows for automation of processes, e.g. running a complete data processing pipeline automatically with new data arriving on a folder that is monitored by an agent. The Workflow system can also deeply integrated in external processes by a command line client that allows for fully unattended execution of workflows. Genedata supports data preservation by means of process protocols, reporting, and archiving. The computer system of Genedata is a workflow management system. Genedata supports multiple users, multiple sites collaboration through its client- server infrastructure. Genedata uses persistent identifiers: CAS-number (for chemical compounds), Ensemble Gene IDs, RefSeq Gene IDs, GUIDs (global unique identifier) (for lab systems). Genedata adopts ontologies; most often used by customers are gene ontology (GO), ontologies provided by NextBio, sequence ontology (SO), taxonomies. The software can be configured to work with any of those ontologies. Data are protected by security features of Oracle, e.g. POSIX from Unix. Transmission is fully encrypted (SSH / HTTPS). rdaeurope@rd-alliance.org | europe.rd-alliance.org 15 16 Legal and Policy Issues Genedata is a closed source company (source code is not made available to the community), but is using open standards for its interfaces and protocols. Algorithms are documented / referenced, and detailed processing reports generated automatically. (pdf reports). Genedata does not adopt restrictions to data-reuse. Limitations /issues Genedata currently does not adopt cloud computing, but this is planned mid- term. Proposed actions (if mature enough) Cluster computing solution will be available by the middle of this year. Community Data Analysis: TNO Community: Netherlands Toxicogenomics Centre (NTC) Interview with TNO as partner of NTC Goals of analysis From interviewing TNO, one of the partners of NTC, we hope to learn more about data organization within a consortium of research institutes and companies. Analysis provided by Maastricht University (UM) Description of the community NTC is a collaboration between universities, research institutes and companies in the Netherlands. TNO is a research institute and one of the partners of NTC. Goals of the community employ toxicogenomics to increase the basic understanding of mechanisms; develop new methods that better chart the risk of chemical compounds; develop alternatives to animal testing; advance collaboration with external partners. toxicological Data types NTC has experimental data from in vitro and in vivo life science experiments (transcriptomics, metabolomics, ...). Metadata formats (for Omics) are SimpleTox (from Array Track) and ISATab. Data flow Exposure to toxic compounds of cell lines / animals is measured at different time points / for different doses of the toxic compound. The different types of data (transcriptomics, metabolomics, ...) are stored in a raw data file. R scripts generate data and graphs for quality control. The data are manually curated after inspection of QC results. Furthermore, meta-data information is manually curated as well. Data are preprocessed and normalized, and finally analyzed with several data mining and statistical tools. After publication, data will be made available to other research projects (online distribution). rdaeurope@rd-alliance.org | europe.rd-alliance.org 16 17 Data organization NTC adopts Data Management Systems: relational database (my SQL). Data exchange The computer environment of NTC is a centralized system with 4 servers, located in Maastricht, that communicate with each other. A backup server is located at TNO. Information about the NTC research projects can be obtained from the NTC website. NTC uses software tools (Metacore, Inguity) to extract data from outside sources. NTC utilizes web services for data exchange with external data sources as well. Data services and Tools For data protection, the following techniques and tools are used: SSH secure shell, user and group authentication, firewall and physical protection. NTC uses persistent identifiers: Entrez Gene ID, KEGG ID, CAS-nr. NTC uses BioPortal, a portal that provides access to commonly used biological ontologies. NTC applies bioinformatics tools for data integration and web services (PubChem, KEGG) for retrieving external information. Data analysis techniques adopted by NTC are, among others: classification, clustering, statistics, functional enrichment, network analysis, quality control, normalization. Data are visualized in heatmaps, PCA, line/scatter plots, networks, boxplots, network visualization, etc. NTC also develops its own tools for data mining, data analysis and data visualization (R scripts). Legal and Policy Issues NTC makes data available through data platforms (GEO, ArrayExpress, ...). Within NTC, data can be re-used on demand. Outside NTC, data will be made available after publication. Users of the data have to collaborate with NTC. Community Data Analysis: ENVRI Goals of analysis From the interview of the ENVRI project we hope to get sophisticated understanding on the data organization within a large environmental community in Europe, which combines a number of various ESFRI research infrastructures, a few non-ESFRI projects and partner organizations. What are common data requirements? What are dissimilarities? What are general and specific data solutions and challenges? The given interview covers 4 key partners of 6: ICOS, LifeWatch, EPOS, and EMSO as described below. Analysis provided by CSC. Description of the community Description The ENVRI project (Common Operations of Environmental Research Infrastructure) is a collaboration conducted within the European Strategy Forum on Research Infrastructures (ESFRI) Environmental Cluster. The ESFRI Environmental research infrastructures involved in ENVRI including: rdaeurope@rd-alliance.org | europe.rd-alliance.org 17 18 ICOS, European distributed infrastructure dedicated to the monitoring of greenhouse gases (GHG) through its atmospheric, ecosystem and ocean networks, http://www.icos-infrastructure.eu/; EURO-Argo, European contribution to Argo, which is a global ocean observing system, http://www.euro-argo.eu/; EISCAT-3D, European new-generation incoherent-scatter research radar for upper atmospheric science, http://www.eiscat3d.se/; LifeWatch, an e-science Infrastructure for biodiversity and ecosystem research, http://www.lifewatch.com/; EPOS, European Research Infrastructure on earthquakes, volcanoes, surface dynamics and tectonics, http://www.epos-eu.org/; EMSO, European network of seafloor observatories for the long-term monitoring of environmental processes related to ecosystems, climate change and geo-hazards, http://www.emso-eu.org/management/. ENVRI also maintains close contact with the other not-directly involved ESFRI Environmental research infrastructures by inviting them for joint meetings. These projects are: IAGOS, Aircraft for global observing system and SIOS, Svalbard arctic Earth observing system. ENVRI IT community provides common policies and technical solutions for the research infrastructures, which involves the following organization partners: Cardiff University, CNRISTI, CNRS (Centre National de la Recherche Scientifique), CSC, EAA (Umweltbundesamt Gmbh), EGI, ESA-ESRIN, University of Edinburgh, and University of Amsterdam. Goals of the community with respect to data The central goal of the ENVRI project is to implement harmonized solutions and to draw up guidelines for the common needs of the environmental ESFRI projects, with a special focus on issues as architectures, metadata frameworks, data discovery in scattered repositories, visualization and data curation. This will to empower the users of the collaborating environmental research infrastructures and enable multidisciplinary scientists to access, study and correlate data from multiple domains for "system level" research. The collaborative effort will ensure that each infrastructure can fully benefit from the integrated new ICT capabilities beyond the project duration by adopting the ENVRI solutions as part of their ESFRI implementation plans. In addition, the result will strengthen the European contributions to GEOSS - the Global Earth Observation System of Systems. All the nine Social Benefit Areas identified and addressed by GEO-GEOSS will take advantage of such approach. Data types EMSO: The EMSO data infrastructure has been conceived to utilize the existing distributed network of data infrastructures in Europe and use the INSPIRE and GEOSS data sharing principles. A number of standards have been set forth that will allow for state-of-the-art transmission and archiving of data with the kinds of metadata recording and interoperability that allow for more straightforward use and communication of data. These standards include the Open Geospatial Consortium (OGC) Sensor Web Enablement (SWE) suite of standards, namely the OGC standards SensorML, Sensor Registry, Catalogue Service for Web (CS-W), Sensor Observation Service (SOS) and Observations and Measurements (O&M). OGC SensorML is an eXtensible Markup Language (XML) for describing sensor systems and processes. Following on progress from EuroSITES and others a SensorML profile is being created that can be stored in a so-called Sensor Registry that will act as a catalogue of each rdaeurope@rd-alliance.org | europe.rd-alliance.org 18 19 EMSO sensor. This dynamic framework can accommodate the diverse array of data and formats used in EMSO, including the addition of delayed mode data. Measurements: a non-exhaustive list of measurements and sensors that can be performed with seafloor observatories is listed below: Water Conductivity, Temperature, Pressure, pH, Eh, alkalinity; Ground motion velocity and acceleration; Earth Gravity acceleration and Magnetic induction field; Geodesy and seafloor deformation (displacement) Gas and dissolved elements concentration; Sound velocity; Heat Flow (temperature). Metadata: EMSO collects metadata on both, the physical sensors and observatories as well as on the data. Observatories are intended to be described by SensorML. Metadata on archived data sets is compatible to ISO19115, DIF or the NetCDF (CF) specification. EPOS: Raw measurements: continuous seismic waveform data in SEED format and corresponding metadata, 1-100'sTB; accelerometric waveform data in ASCII. Measurements / Quality Control data: Power Spectral Density: PSD of the background noise as function of time, for selected frequencies. PSD database (PQLX) and Probability Density Function (PDF) representation (PQLX database). Magnitude: histograms for magnitude differences between station magnitude and VEBSN magnitude. Time residuals: time residuals distribution for each station. Metadata: metadata definitions are currently an important topic of discussion. Within seismology a task force will be established to define and store the concepts and the vocabulary terms for the metadata items. Dataless SEED is the current international standard format to describe instrument characteristics. (Derivative XML formats are also in use, but common agreement has not been reached yet). The main requirements or the next phase of the metadata definition can be listed as following (EUDAT initiative): A simple „flat‟ metadata standard for discovery; (flat metadata means it is a single record with attributes rather than a group of linked records each with attributes and with relationships between the records); A structured (linked entity) standard for context (relating the dataset to provenance, purpose, environment in which generated etc); Detailed metadata standards for each kind of data to be co-processed; The following standards appropriate to support such model: Discovery: DC; Contextual: CERIF (Common European Research Information Format ) or ISO 19115; Detailed: Individual standards depending on type of dataset; for research datasets from large-scale facilities CSMD (e.g http://www.ijdc.net/index.php/ijdc/article/view/149, see also PaNData, http://www.pan-data.eu/PANDATA_– rdaeurope@rd-alliance.org | europe.rd-alliance.org 19 20 _Photon_and_Neutron_Data_Infrastructure), for geospatial datasets INSPIRE 1, http://inspire.jrc.ec.europa.eu/, http://en.wikipedia.org/wiki/INSPIRE (as in ENVRI). Derived/processed data, publications, software: Earthquake catalogues represented in many different formats, from text based to XML. ICOS: Data hierarchy of ICOS differs between its two Thematic Centers (Atmospheric and Ecosystem). Data hierarchy in ICOS Atmospheric Thematic Center (ATC) is divided into 4 levels defined as: Level-0: Raw data (e.g. current, voltages) produced by each instrument; Level-1: Parameter expressed in geophysical units. For example it can be GHG concentrations (e.g. ppm-CO2). Level 1 is also dived into two levels: Level 1.a: Rapid delivery data (NRT, 24hr) and level 1.b: Long term validated data; Level-2: Elaborated products. For GHG concentrations it can be, e.g., gap-filling, selection, etc. Level-3: Added value products (to be defined with AS PIs). Metadata are provided by PIs via a graphical applications developed at ATC. Raw data are daily transferred to ATC where data are automatically processed. Those raw dataset are mainly ASCII files, depending on considered instrument. A specialized processing chain is dedicated for each type of instrument deployed in ICOS Atmospheric Station. The process involves the transformation of raw data (Level 0) to upper level product. A level 1 ICOS atmospheric station is continuously measuring 18 parameters (among them greenhouse gases, meteorological parameters and planetary boundary layer height). Most data are continuous measurements. About 400 Mo by level 1 ICOS Atmospheric Station is daily uploaded to ATC. Considering that ICOS Atmospheric network will comprise about 50 atmospheric observatories, amount of data produced is estimated to be around 20Go/day, i.e. 7.3To/yr. Note that is an upper bound value since all stations are not going to be labeled as level-1 ICOS atmospheric station. Data catalogue of produced datasets is not yet automatically available but is intended to be. Data hierarchy in ICOS Ecosystem Thematic Center is divided into 5 levels defined as: Level-0: Raw data; Level-1: First set of corrections applied to the raw data; Level-2: Consolidated half-hourly fluxes; Level-3: Standardized QAQC and filtering applied to the half hourly data; Level 4: Data gap filled and aggregated at different resolutions; Level 5: Derived variables calculated) data products. The data collected at the Ecosystem Sites are raw data at 10 Hz time resolution. These data need a first processing step to calculate greenhouse gas fluxes with typical time resolution of 30 min. These fluxes are further corrected, filtered, gap-filled where necessary, and processed to retrieve additional variables. LifeWatch: Raw measurements: These are mostly generated by sensors, resulting in data such as organism presence/absence/abundance; species identification; or physiological data (i.e. plant respiration). Measurements: Long-term monitoring programmes are managed by established networks for terrestrial and for marine environments. Data are: species composition, biomass, phenology, decomposition, etc. Observations: (mostly human) observations of species presence (identification, date/time, spatial coordinates). rdaeurope@rd-alliance.org | europe.rd-alliance.org 20 21 Derived/processed data, publications, software: these are input data for other users. Data flow By examining computational characteristics of participating ESFRI environmental research infrastructures, we have identified 5 common subsystems: Data Acquisition, Data Curation, Data Access, Data Processing and Community Support. Typical data lifecycle spans over five subsystems. This lifecycle begins with the acquisition of raw data from a network of integrated data collecting instruments (seismographs, weather stations, robotic buoys, human observations, etc.) which is then preprocessed and curated within a number of data stores belonging to an infrastructure or one of its delegate infrastructures. This data is then made accessible to authorised requests by parties‟ out with the infrastructure, as well as to services within the infrastructure. This results in a natural partitioning into data acquisition, curation and access. In addition, data can be extracted from parts of the infrastructure and made subject to data processing, the results of which can then be situated again within the infrastructure. Finally, the community support subsystem provides tools and services required to handle data outside of the core infrastructure and reintegrate it when necessary. Data organization Typical granularity of a data set of EMSO is e.g. for given period of time (monthly, duration of an experiment), or for a given instrumentation (see examples at PANGAEA or MOIST). Currently the following distinction is made among the data levels within EPOS: L0 – raw data; L1 – QC data; L2 – filtered data; L3 – research level pre-processed data; L4 – research product Organization of ICOS ATC and ETC data is based on levels described in the first point (type of data). ICOS Level 3 products defined as “Added value products” is under consideration with PIs, but will include dataset resulting from aggregation of multiple lower levels ICOS products. LifeWatch data are of very different kinds: species information, species distributions, species abundance, biomass and distributions, species DNA sequences, genes, earth observation data (canopy cover etc), species compositions, age distributions, radar data, etc. Data exchange Within EPOS, the data are received at the data centers in real time, through dedicated TCP/UDP connections to the sensors, adopting a widely known application level protocol (SeedLink). For ICOS, data are daily uploaded from ICOS Atmospheric station on a dedicated ftp server at ICOS ATC. Note that data can be exceptionally provided via attached document in an email. ATC Raw data are automatically ingested into a MySQL database, and then processed. ICOS Ecosystem sites submit their raw data monthly to the ICOS ETC. In addition, preliminary half hourly fluxes and meteorological data will be transferred automatically to the ETC in Near Real Time (one day). rdaeurope@rd-alliance.org | europe.rd-alliance.org 21 22 LifeWatch data are generated by various external data providers at the European and global scale. LifeWatch is deploying (shared) data for analysis and modeling. Data reside at the external data providers, and LifeWatch data catalogues assist users in data discovery. Data services and Tools Most of the projects are still in their construction phase and their services are not yet fully operable. Service partially in operation and/or under implementation within EMSO: PANGAEA OAI-PMH for ESONET data in EMSO sites: harvesting test, integration into ENVRI metadata catalogue etc; PANGAEA GeoRSS : Embedding GeoRSS feed; Ifremer SOS for EUROSITES oceanographic data in EMSO sites: getCapabilities; getObservation, check O&M format; PANGAEA SOS for INGV data in EMSO sites (via MOIST: moist.rm.ingv.it); getCapabilities, getObservation, check O&M format!! MOIST OpenSearch for INGV data and metadata in EMSO sites: Data and metadata search according to time or space or parameter; Common NetCDF metadata extraction and transformation service: Metadata extraction; MOIST OAI-PMH for harvesting INGV data and metadata in EMSO sites: Data and metadata harvesting. LifeWatch is an e-Science infrastructure facilitating biodiversity and ecosystem research with state- of- the-art ICT services for analysis and modelling in the context of a systems approach to tackle biodiversity complexity. LifeWatch is at the beginning of its Construction Phase and comprehensive realizations of services have not yet been done. Web services are intended and pilot implementation of these is being undertaken within the Biodiversity Virtual e-Laboratory (BioVeL) project. There are six different levels of services within LifeWatch: Communication services with integration, transformation, messaging, encoding and transfer services; Information management services: with data access, thematic data access, annotation, identification, discovery, mediation and user management services; Processing services with spatial processing, analytical and modeling, taxonomic processing, visaulisation, thematic processing, metadata and integration services; Human interaction services with portrayal, thematic interaction, interaction, personalization and collaboration services; Workflow services with orchestration services; and System management services with security, quality evaluation and provenance services such as monitoring, service management and transaction services. For more details see the LifeWatch Reference Model documentation. Once the metadata structure will be defined for EPOS, the searching capabilities will be implemented adopting standard interfaces, e.g. OGC web services. The carbon portal of ICOS will allow for data search across the different ICOS databases. Data search restricted to geographical areas or time periods, of similar nature than the one rdaeurope@rd-alliance.org | europe.rd-alliance.org 22 23 developed on the INSPIRE GEOPORTAL, will be implemented. The CP will also act as a platform to offer access to higher level data product and fluxes. By incorporating state-of-the art Information Retrieval methodologies, LifeWatch will enable scientists to query across datasets and discover statistically significant patterns within all available information. This form of Information Retrieval assumes a model of the world based on the available information in the datasets, and applies statistical methods to reveal patterns in this model. LifeWatch recognizes the need for well-defined semantics and uniformity in datasets and stimulates the practice by promoting standards and protocols. But at the same time it realizes that this is an ambition only feasible on the long-term and thus needs a pragmatic approach to suit the needs of scientists on the shorter term. Information Retrieval methodologies, already well-established in other scientific fields, will supply this pragmatism and enable LifeWatch to build intelligent query interfaces even without structured data. Within EPOS, VERCE will lay the basis for a transformative development in data exploitation and modeling capabilities of the earthquake and seismology research community in Europe and consequently have a significant impact on data mining and analysis in research. LifeWatch services include custom-made toolboxes for various user (research) areas, consisting of applications to be combined to preferred workflows. Such applications will cover sets of related algorithms. Legal and Policy Issues Most of the projects follow the open data sharing policy. The vision of EMSO is to allow scientists all over the world to access observatories data following an open access model. Within EPOS, EIDA data and Earthquake parameters are generally open and free to use. Few restrictions are applied on few seismic networks and the access is regulated depending on email based authentication/authorization. The ICOS data will be accessible through a license with full and open access. No particular restriction in the access and eventual use of the data is anticipated, expected the inability to redistribute the data. Acknowledgement of ICOS and traceability of the data will be sought in a specific, way (e.g. DOI of dataset). A large part of relevant data and resources are generated using public funding from national and international sources. LifeWatch is following the appropriate European policies, such as: The European Research Council (ERC) requirement that data and knowledge generated by public money should also become available in the public domain, http://erc.europa.eu/pdf/ScC_Guidelines_Open_Access_revised_Dec07_FINAL.pdf . The European Commission’s open access pilot mandate in 2008, requiring that the published results of European-funded research in certain areas be made openly available. For publications, initiatives such as Dryad instigated by publishers and the Open Access Infrastructure for Research in Europe (OpenAIRE). The private sector may deploy their data in the LifeWatch infrastructure. A special company will be established to manage such commercial contracts. rdaeurope@rd-alliance.org | europe.rd-alliance.org 23 24 Limitations /issues When software code can also regarded as data, there is the unresolved issue of identities, provenance and interoperability of software codes (for example as components in a workflow); Identity of concepts; Streaming data analytics (computation) of parallel real-time data streams; Interoperability between different research infrastructures e.g., how to support integrated data discovery and access of heterogeneous scientific data. Proposed actions (if mature enough) No efforts yet; See example of Global (species) Names Architecture: http://www.globalnames.org/; Considered by the EUDAT project; In ENVRI, OpenSearch technology is used to create a web data portal which allows users to discover and access data residing at the different federated Digital Repositories (DR) sites on the basis of personal search criteria. Space for general remarks We have realized there is an urgent need of developing a Common Reference Model for the community. A Reference Model is a standard and an ontological framework for the description and characterization of computational and storage infrastructures in order to achieve seamless interoperability between the heterogeneous resources of different infrastructures. Such a Reference Model can serve the following purpose: To provide a way for structuring thinking which helps the community to reach a common vision; To provide a common language which can be used to communicate concepts concisely; To help discover existing solutions to common problems; To provide a framework into which different functional components of research infrastructures can be placed, in order to draw comparisons and identify missing functionality. Only by adopting a good reference model can the community secure interoperability between infrastructures, enable reuse, share resources and experiences, and avoid unnecessary duplication of effort. Community Data Analysis: Svali Goals of analysis From the interview of the SVALI consortium we hope to learn more about data organization within well standardized and organized Nordic scientific community. rdaeurope@rd-alliance.org | europe.rd-alliance.org 24 25 Analysis provided by CSC. Description of the community Description The SVALI consortium (Stability and Variations of Arctic Land Ice) Initiative (TRI, www.topforskningsinitiatived.org) Top-level Research NCoEs DEFROST experience and access to other RIs. The partners are working with focus on the Arctic and Sub-Arctic. Goals of the community with respect to data TRI NCoE ICCC aims at a joint Nordic contribution in cryospheric studies to solve one of the most important global climate change research challenges. The programme integrates studies on stability of glaciers, atmospheric chemistry and biogeochemistry. The SVALI collaboration is based on three pillars: 1) common analysis, interpretation and reporting of changes in the cryosphere in the North Atlantic area; 2) common platform for graduate studies and postgraduate research work between the main research institutions and universities involved in cryospheric studies based on exchange of students and researchers, a common pool of observational data and a joint programme for organization and for obtaining support for future cryospheric research, and 3) using NCoE ICCC as a vehicle for wider international collaboration within cryosphere research in the Nordic countries. The objective of NCoE ICCC data management is to ensure the security, accessibility and free exchange of relevant data that both support current research and future use of the data. The main goals of the SVALI community with respect to data are to facilitate open access to SVALI results for research within and outside of SVALI, to ensure safe storage and availability of relevant data beyond the SVALI project period and efficient exchange of data between SVALI partners in collaborative research efforts. Data Types SVALI operates with observational and experimental data, i.e. collected by measurement and computational data produced by simulations. The main data types are (1) remote sensing data of various types (SPOT5, Aster, ICESat, Cryosat-2, ERS, Tandem-X, GRACE, ...) as well as data from airborne lidar surveys, (2) Field data such as mass balance measurements, GPS measurements, data from meteorological stations on glaciers, etc., (3) model simulation results (data from Earth system models (EC-Earth), meteorological models (WRF, HIRHAM5), ice flow models (Elmer, PISM) and mass balance models. These data are typically stored in special format that is appropriate for each type, such as various image formats for remote sensing data, netCDF (modelling results), grib (EC-Earth and HIRLAM5), las (point cloud lidar data), etc. Data Flow Remote sensing data are obtained from international space agencies and data centres. This often involves internet data access ports with online application forms for gaining access to the data. Field data are gathered by individual project partners and simulation results are similarly created by individual partners. In some cases, simulation results are created at computer centres (CSC, ECMWF), pre- processed there and preprocessed data are transferred for further rdaeurope@rd-alliance.org | europe.rd-alliance.org 25 26 analysis to the computer system of the respective partner. Simulations (e.g. EC-Earth) often create vast amounts of data in temporary storage areas that are deleted after further processing where e.g. monthly averages are computed from data files with higher temporal resolution. The raw model results may be created in specialized format, e.g. grib, and reformatted into more general purpose format, e.g. netCDF, in the preprocessing. After processing, data may be made available to other project partners over the internet or submitted to international data archives for storage or further analysis. Data Organization NCoE ICCC data are those data generated during the duration of the three individual NCoEs within NCoE ICCC (October 2010 – September 2015) through work packages that are organised as part of the individual NCoEs. Data generated by simulation models or gathered in the field measurement campaigns as a SVALI activity, are freely available to all SVALI participants and to other scientists as appropriate on the shortest feasible timescale in accordance with the TRI/ICCC data policy. To facilitate SVALI data storage and sharing, data providers are requested to submit a data description (or metadata) by filling a corresponding template and send it to the dissemination team at GEUS, Denmark. The metadata will be available at the SVALI website (http://ncoe-svali.org/data). Data of international importance collected within SVALI should be stored for longer term than the duration of the SVALI NCoE and will be submitted to appropriate data centers, depending on the nature of the data. Existing data centers, such as the National Snow & Ice Data Center (www.nsidc.org) or the World Glacier Monitoring Service (www.geo.uzh.ch/microsite/wgms) will be used, rather than the SVALI NCoE creating own storage of data. Formal contact has been made with GCW (WMO programme Global Cryosphere Watch) as well. Some SVALI data (not suitable for storage in NSIDC and WGMS) will be uploaded to the forthcoming GCW data portal (http://gcwdemo.met.no) together with appropriate metadata. This will ensure long-term storage of these data beyond the 5-year TRI period. If this does not work out, other options for ensuring long-term storage will be explored before the end of the project period. Data that are primarily of importance within the SVALI project and alpha- and beta- versions of data sets that need to be shared between project partners will be stored on externally accessible servers as appropriate with metadata descriptions available on the SVALI web. Data Exchange Data are mainly exchanged between partners through the internet (ftp, web-based download from internet servers). Some partners and collaborating institutes have or are developing internet data download centres that provide access to data and maintain a log of the downloads (who is downloading, for what purpose, etc.). Some SVALI created simulation data are available as part of international databases for providing access to model simulation results. As mentioned above, it is the policy of SVALI that data are as far as possible to be submitted to international archives where they are openly available for research. Data Services and Tools The SVALI community uses a variety of software tools for storing, exchanging, processing, analyzing, visualizing data. Software and software packages such as grib, netCDF, Matlab, R, python, FERRET, ERDAS, netCDF tools (nco, ncBrowse, netCDF read/write libraries for analysis software) are among the the most important software packages. Data are mainly exchanged between partners over the internet (ftp, sftp, web-based download). Legal and Policy Issues rdaeurope@rd-alliance.org | europe.rd-alliance.org 26 27 The SVALI NCoE adheres to an Open Data Policy. The TRI/ICCC Data Policy, which applies to the SVALI NCoE as well as to other TRI/ICCC NCoE, is based on the existing “International Polar Year Data Policy”. The aim of the data policy, as for the IPY policy, is to provide a framework for data to be handled in a consistent manner, and to strike a balance between the rights of investigators and the need for widespread access through the free and unrestricted sharing and exchange of both data and metadata. The policy is compatible with the data principles of the Top-level Research Initiative (TRI, “http://www.toppforskningsinitiativet.org/en”). The data policy is reviewed annually by the Steering Group and any updates will be formally signed by the Project Leader to record their formal adoption and for issue controlling. Limitations/issues Some particularly important data archives for NCoE ICCC are the National Snow and Ice Data Center (NSIDC, http://nsidc.org), World Glacier Monitoring Service (www.geo.uzh.ch/microsite/- wgms), archives related to WCRP CliC and the newly established GCW. It must be recognized that data preservation and access should not be afterthoughts and need to be considered while data collection plans are developed. A subset of data both generated and used by NCoE ICCC needs a specialized policy and access considerations, because they are legitimately restricted in some way. Access to these data may for example be restricted because there may be intellectual property issues. It is the overall aim of NCoE ICCC that data are as freely available as possible within the constraints provided by such legitimate restrictions. Proposed actions (if mature enough) Not applicable Space for general remarks None Community Data Analysis: EISCAT 3D Goals of analysis From the interview of the EISCAT_3D research infrastructure we hope to learn on different aspects of the data organization (general, technical, legal) of the project inside specific international scientific community starting its preparatory phase and find out what are the requirements and challenges facing the high attitude project in the field of data management. Analysis provided by CSC. Description of the community Description EISCAT_3D (European 3D Imaging radar for atmospheric and geospace research, E3D) will be a world-leading international research infrastructure using the incoherent scatter technique to study the atmosphere in the Fenno-Scandinavian Arctic and to investigate how the Earth's atmosphere is coupled to space. E3D is led by Swedish EISCAT Scientific rdaeurope@rd-alliance.org | europe.rd-alliance.org 27 28 Association. Current participants involved are EISCAT partners [China, Finland, Japan, Norway, Sweden, and UK], associated partners [France, Russia, Ukraine] and EISCAT user communities. Goals of the community with respect to data EISCAT_3D is designed for continuous operation, capable of imaging an extended spatial area over northern Scandinavia with multiple beams, interferometric capabilities for small-scale imaging and with real-time access to the extensive data. The goal of the community is continuous measurement of the space environment – atmosphere coupling at the southern edges of the polar vortex and the aurora oval. E3D will be a key facility for various researches and operational areas including environmental monitoring, space plasma physics, solar system science and space situational awareness. In addition, EISCAT_3D will provide a platform to develop new applications in radar technology, experiment design and data analysis. Data Types EISCAT operates initially with raw observational data. The following data types are currently in use: EISCAT raw data (Matlab binary); analysed data (Matlab binary, ASCII, HDF5, etc.); CDF format for metadata and analysed data; KMZ format for analysed data; ps, pdf, png for summary plots. EISCAT-3D will operate with a huge volume of data. The structure of these data is considered and metadata is expected to be in accordance with standards. Data Flow The EISCAT_3D facilities will comprise one core site and at least four distant sites equipped with antenna arrays, supporting instruments, platforms for movable equipment and high data rate internet connections. The key part of the core site is a phased-array transmit/receive (TX/RX) system consisting of roughly 10,000 – 16,000 elements and other state-of-the-art signal processing and beam-forming instruments. Each antenna produces 2 x 32bit/sample x 30 Msamples/s (= 2Gbit/s). At 25% duty cycle this is 5 Tbit/day and the data rate of a 16,000 antenna array would be about 80 Pbit/day. Antenna group computes a number of beams from a small number of antennas. The antenna group of about 100 antennas forms a limited number of polarized beams at a selected limited bandwidth I/O. These beam- formed data are stored in a ring buffer for a relatively long duration (hours to days). As the full arrayproduces a raw data rate which is too large to be archived, one has to limit the archived data rates so that e.g. 160 antenna groups form 100 beams with total maximum of 20 Gbit/s data to be stored in archive. At least one set of time-integrated correlated data will be calculated from each set of beam-formed data, and permanently stored in a Web accessible master archive. One or several analyzed data sets will be permanently stored corresponding to each set of correlated data. Next, for further offline work these data shall be transferred from on-site archives to HPC. All data should exist at least at two independent sites, archive and datawarehouse, so that archive and data warehouse works even if one site is offline Well- functioning networks with minimum of 10Gbit/s are required and the starting archiving rate would be of the order of 50PB/year. Data Organization Currently, EISCAT archive is small and about 60TB. This is because EISCAT was not archiving sampled raw data from the start of operations in year 1981. Instead, what often is called EISCAT raw data is in fact correlated data samples, the so called auto-correlation function estimates, which are organized and stored for further analysis as lag-profiles, altitude profiles of different time lags of the auto-correlation function of the signal. The final analyzed EISCAT rdaeurope@rd-alliance.org | europe.rd-alliance.org 28 29 data, physical ionospheric and atmospheric parameters, such as e.g. Electron density and temperature as function of time and altitude etc., are available and organized in the Madrigal DB. Madrigal is an upper atmospheric science DB, used by research groups world-wide and originally specially designed for incoherent scatter radar (ISR) data. Madrigal is a distributed DB, where data at each Madrigal site is locally controlled and accumulated, but shared metadata between Madrigal sites allow searching of all Madrigal sites at once. Madrigal is built so that it also can handle future EISCAT_3D data, as it is already handling data from the US and Canadian phased-array radars, PFISR (Poker Flat ISR) and RISR (Resolute Bay ISR). However, further development of volumetric data products is also envisaged. This development will be of benefit for the whole global ISR community. Data Exchange The future system will generate very large volumes of data. An efficient archive and data warehouse shall be deployed during the construction phase using existing e-infrastructures in Northern Scandinavia and synergies with resource centers. At least two independent sites shall support all data to be accessible, backed up and secured. The expected stored data volume in the initial phase of operation is of the order of more than 1000 TB per year. Data Services and Tools Currently, the EISCAT community uses: Madrigal DB, IUGONET Metadata DB, UDAs – IUGONET Data Analysis SW, Dagik visualization tool, CEF (Conjunction Event Finder) web tool for seamlessly browsing quick-look data. Users may also do re-analysis of the data. The basic analysis software GUISDAP is available for platforms running Matlab and there is also a web interface to use GUISDAP if simplified analysis control is all that is needed, which is often the case for standard radar experiments. Future E3D tools are under consideration. In the Preparatory Phase of EISCAT_3D, one is concentrating the development to solutions for the analysis of the data, which would use the latest ISR data analysis strategies. Algorithms and SW modules are being developed for parallelization of the computational tasks both in SW-based beam forming and analysis of multi-beam data and imaging applications. Resources for development of high-level end user data services and visualization tools are not included in the Preparatory Phase. Such work is assumed to be the task of the user communities and their networking efforts with communities who use similarly structured scientific data. Legal and Policy Issues EISCAT has established data policies and procedures for user access. Those will be adapted to the new system keeping in mind the importance that the project places on attracting new users. The new products should target the following four groups of users: 1) experienced EISCAT users; 2) new users attracted by the enhanced conventional capabilities and/or of the new E3D capabilities; and 3) environmental and space weather modelers and service providers. Occasional users interested in E3D for short-duration research projects or as source of supporting data. Data are open and shared internationally by corresponding national research organizations with 1 year exclusive right.Long storage of data is required, both because the nature of the geophysical data as a record of space weather and solar- terrestrial relations with minimum time-scales covering several solar cycles (each 11 years) is relevant to climate change research, as well as due to the fact that re-analysis of raw data would bring in possibly new interpretations, improve results and reveal unforeseen natural processes. Minimum preservation time of data would be 30-40 years. rdaeurope@rd-alliance.org | europe.rd-alliance.org 29 30 The proposed new EISCAT agreement, which would cover EISCAT_3D, considers 3 types of participation and financial contributions to the research infrastructure. All these have different implications to data policy. The in–kind Core Science investment is fully relevant to EISCAT core science and in line with the scientific and strategic priorities. Operational costs are then fully met by EISCAT. Open access to data and compliance with the EISCAT data and user access policy is required for such contributions. In-kind Mix of Core and Non-Core Science investment is partly relevant to EISCAT core science, but not fully in line with its scientific and strategic priorities. Operational costs are divided between EISCAT and contributing Associate or Affiliate in proportion to contribution to EISCAT core science and strategy. Open access to data is required in this case, too. Hosting contributions are using an EISCAT site and infrastructure but there is no value to EISCAT core science. Operational costs are then fully met by the contributing Associate, Affiliate or 3rd party. Open access to data is encouraged but not required. Limitations/issues Towards its 1st stage by 2018, EISCAT_3D needs moderate level of archive size about 1PB/year and more HPC capacity (1 Pflop/run) and storage 1 PB. By the next stage in 2023, EISCAT archive is expected to be about 50 PB/year, and HPC performance will up to 1000 Pflop/run. The key issue is transferring data from sites for processing, and fast internet connection is strictly required. Proposed actions (if mature enough) The E3D is now in the Preparatory Phase. Its aims to ensure that E3D project will reach a sufficient level of maturity with respect to technical, legal and financial issues so that the construction of the next generation E3D radar system can begin immediately after the conclusion of the phase. A new EISCAT agreement will be finalized in November 2013. The E3D preliminary design review is planned for October 2014. The EISCAT_3D Preparatory Phase currently works under 14 work packages that cover different aspects of the advanced infrastructure. Taking into accounts E3D needs in HPC and storage capacity, collaboration with EUDAT could be considered. The present EISCAT is already fully integrated in the global network of incoherent scatter radars. E3D is an environmental RI on the EU ESFRI roadmap. Space for general remarks The E3D preparatory phase (October 2010-September 2014) is funded by EC under the call FP7- INFRASTRUCTURES-2010-1 “Construction of new infrastructures: providing catalytic and leveraging support for the construction of new research infrastructures”. The EISCAT implementation time line (2014-2021) incorporates a smooth transition from preparation to implementation in 2014, provided that sufficient funds are allocated, subsequently construction in 2016, and the first operation in 2018. Community Data Analysis: Public Sector Information ENGAGE Goals of analysis Open Public Sector Information (PSI) can be anything varying from election results, statistics from population, unemployment, earnings and transportation to fire incidents, criminal records and illegal immigrants. As the PSI files are mostly unstructured and in non-machine rdaeurope@rd-alliance.org | europe.rd-alliance.org 30 31 processable formats(e.g. pdf), it is interesting to see how the ENGAGE curation community works and what tools it uses to make them more structured and what kind of metadata approach is needed to treat such data. Analysis provided by ATHENA. Description of the community The ENGAGE community consists of researchers, innovators, government employees, software developers, media journalists and citizens who create, improve, use, analyze, combine, report on, visualize open data. Goals of the community The goal of the ENGAGE community is to make Public Sector Information (PSI) data openly available with data curation and cleaning facilities, improved metadata, appropriate analysis/visualization tools and portal access. Context: Current practices - achievements and limitations: The ENGAGE platform currently links to original PSI data and derived / curated datasets created, maintained and extended by users (researchers, citizens, journalists, computer specialists) in a collaborative environment. Therefore the ENGAGE is a research / data curation community platform with focus on the Social Sciences and Humanities domain. The vision of the ENGAGE infrastructure is to extract, highlight and enhance the re-use value of PSI data. This can be achieved by moving slowly from low-structured, isolated, difficult to find PSI data to high-structured, easy to link, easy to process datasets through crowd-sourcing. Data types Open data covers almost all research disciplines but currently available datasets are mainly in social sciences, meteorology and transport. Right now the majority are in pdf format with little or no metadata. Next comes .csv or .xls files again with little or no metadata. Around 4% has CKAN or DC as metadata and stored in tables (including Excel) or RDF triples. The rest are mainly in tabular format (relational) although commonly as files rather than databases. Data flow Datasets metadata are harvested or uploaded to the ENGAGE platform where the metadata is enhanced (some automation, dominantly manual) and made openly available (subject to any rights management / licensing restrictions). Data organization ENGAGE stores metadata using the CERIF (Common European Research Information Format – an EU Recommendation to Member States). In addition the ENGAGE platform provides a single point of access to Public Sector Information. Users are able to extend / revise these datasets with a description and type of the extension (e.g. Conversion to other format, Data Enrichment, Metadata enrichment, Snapshots of real-time data, datasets mashups). Users are able to track the entire history of the extensions up to the original dataset through a graph-based diagram of the revisions. rdaeurope@rd-alliance.org | europe.rd-alliance.org 31 32 Data exchange The ENGAGE portal provides metadata and pointers to the open dataset(s). It therefore facilitates interoperation and dataset co-use / mashup. A user is able to upload a new dataset or extend an existing one. The user is allowed to set a maintaining group for this dataset and thus giving managing rights to all the members of this particular group. Data services and Tools The tools cover data upload/download, metadata improvement, data cleaning, analysis and visualization. Also data community/social networking tools. In detail the ENGAGE platform supports the following tools: Browse / search for datasets through faceted search Upload / Bulk Upload datasets Download datasets Request datasets Extend / Revise Datasets Visualise Datasets (Core visualization for tabular datasets - Creating chart based visualizations, Creating map based visualizations, integrating visualizations from external engines – e.g. Many Eyes) ENGAGE plug-in for Open Refine Clustering Analysis (K-means clustering) Manage / share related items (Publications, Web applications, APIs related to the dataset) SPARQL endpoint Restful ENGAGE API (JSON format) ENGAGE Wiki Dataset rating and commenting system Legal and Policy Issues ENGAGE strives for a common CC-BY license but records other licensing regimes. Limitations /issues The only limitations are the availability of open data and – particularly – the poor quality of existing metadata. rdaeurope@rd-alliance.org | europe.rd-alliance.org 32 33 Proposed actions (if mature enough) We are looking at automated metadata improvement while providing facilities for human metadata improvement. Community Data Analysis: ESPAS Goals of analysis As the ESPAS community receives data from a variety of sources of different formats and with different practices, it is interesting to observe how such data is treated and how a common denominator may be achieved. Analysis provided by ATHENA. Description of the community Description The ESPAS community consists of researchers in the area of near earth space, i.e. the upper layers of the atmosphere (ionosphere, lower magnetosphere). Goals of the community ESPAS aims at building the data e-Infrastructure necessary to support the access to observations, the modeling and the prediction of the near-Earth space environment (extending from the Earth's atmosphere up to the outer radiation belts). Data types ESPAS data are outputted from a wide range of available instruments to monitor the nearEarth space: both ground-based ones (such as coherent scatter radar, incoherent scatter radar, GNSS receivers, beacon, ionosondes, oblique sounding, magnetometers, riometers, Neutron monitor, Fabry-Perot interferometers) and space-based ones (ELF/VLF wave experiments, radio spectrometers, Langmuir probes, high energy particle spectrometer, electric and magnetic sensors, energy particle sensors, radio occultation experiments, radio plasma imagers, coronal imagers and EUV radiometers, coronographs). Moreover, there is data derived from models, such as the Physics-based plasmaspheric kinetic model, EDAM and CMAT2. The ESPAS data files outputted from the instruments/models are either numerical data or images that describe the observations. There are various file formats available for each category of data (numerical, images) that are supported from ESPAS data providers. So, for the numerical data the available file types are the following: text files (general, CEF, SAO, SAO.XML, DVL and RINEX) and binary files (CDF, GDF, HDF5, MATLAB (.mat), netCDF, PDF, RDF, RSF and SBF). For image files the following file formats are available: FITS, GIF, JPG, PDF and PNG. The variety of the file formats corresponds to the large variety of the origins of the data (data from different instruments, different experiments and different organizations measuring different physical properties) and it is taken into consideration for the development of ESPAS system. Data flow rdaeurope@rd-alliance.org | europe.rd-alliance.org 33 34 Datasets metadata are harvested from the data providers via the ESPAS platform where the metadata is enhanced and made openly available (subject to any rights management / licensing restrictions). The datasets themselves will be harvested via the ESPAS platform only when needed for the computation of algorithms and models (in the general case only metadata is harvested and not the actual dataset). Data organization ESPAS data providers do not have a common approach to metadata creation, data organization and open access to it. ESPAS has defined a common metadata format based on OGC (Open Geospatial Consortium) observation and measurements standard, http://www.opengeospatial.org/standards/om). Moreover, ESPAS has defined and maintains vocabularies (ontologies) for scientific terms used in the metadata (e.g. http://ontology.espasfp7.eu/observedProperty). Data exchange ESPAS has decided the use of OGC Catalogue Service API as a means to exchange metadata between the ESPAS platform and the data providers, and also between the external users and the ESPAS platform. OGC Sensor Service is also used for the exchange of actual data. Data services and Tools The tools cover data upload/download, metadata improvement, data cleaning, analysis, algorithm execution and visualization. Legal and Policy Issues Each data provider has a different data access policy (from open access to closed data). ESPAS tries to handle all different cases and also strives to adopt a more open approach. Limitations /issues The only limitations are the availability of open data and the wide variety of data formats. Proposed actions (if mature enough) In order to homogenize the file formats, the data providers are encouraged to implement the OGC Sensor Service API that defines a common data format. Math Community: combinatorics On-line databases : from L-functions to Edinburgh, 21-25 January 2013 Françoise GENOVA, Final version, 13 March 2013 The workshop The workshop1 sponsored by the American Institute of Mathematics, the International Centre for Mathematical Science (Edinburgh), and the National Science Foundation, was devoted to 1 http://www.aimath.org/ARCC/workshops/onlinedata.html rdaeurope@rd-alliance.org | europe.rd-alliance.org 34 35 the development of new software tools for handling mathematical databases. These tools will assist mathematicians in the integration, display, distribution, maintenance and investigation of mathematical data, particularly in the context of the computer algebra system Sage. F. Genova was invited to present data centres in astronomy and their development, in particular the International Virtual Observatory. This note is based on information gathered during the workshop and on discussions with the participants. Context The starting point is the Sage free open-source mathematics software system 2, a community effort which aims at creating a viable free open source alternative to the commonly used software Magma, Maple, Mathematica and Matlab. “Sage is built out of nearly 100 open-source packages and features a unified interface. Sage can be used to study elementary and advanced, pure and applied mathematics. This includes a huge range of mathematics, including basic algebra, calculus, elementary to very advanced number theory, cryptography, numerical computation, commutative algebra, group theory, combinatorics, graph theory, exact linear algebra and much more. It combines various software packages and seamlessly integrates their functionality into a common experience. It is well-suited for education and research. ” There are currently 250 contributors in 170 different places from all around the world3. Sage is a large ecosystem, and the workshop gathered two communities engaged in addedvalue efforts on two different but nearby topics, number theory and combinatorics, looking for possible convergence of their efforts: The LMFDB project, which aims to gather data in number theory, as relevant to the study of L-functions The Sage-combinat project and related projects, concerned with combinatorial problems As it will appear in the following, LMFDB and Sage-combinat are both strongly related to Sage but with different types of relations. LMFDB, the L-functions and modular form database “LMFDB4 is the database of L-functions5, modular forms, and related objects. It intends to be a modern handbook including tables, formulas, links, and references for L-functions http://www.sagemath.org/ http://www.sagemath.org/development-map.html 4 http://www.lmfdb.org/ 5 Wikipedia definition of L-Functions : “In mathematics, an L-function is a meromorphic function on the complex plane, associated to one out of several categories of mathematical objects. An L-series is a power series, usually convergent on a half-plane, that may give rise to an L-function via analytic continuation. ”. LMFDM definition : “By an L-function, we generally mean a Dirichlet series with a functional equation and an Euler product.”which can be explored from http://www.lmfdb.org/knowledge/show/lfunction.definition. 2 3 rdaeurope@rd-alliance.org | europe.rd-alliance.org 35 36 and their underlying objects.” It is thus a handbook for a special class of functions, with lots of connections to basic analysis and other related objects 6 . The information is very well structured, and the database has rich searching functionalities. It implements elements coming from Sage, but also other information. It aims at being as complete as possible, and its implementation has triggered research to fill gaps. It includes an “Encyclopedia” technically based on knowls 7 , which dynamically includes relevant, supplementary information in web pages. Each sub-domain is coordinated by a specialist who is a member of the editorial committee. The target audience is other number theorists and students, with the hope to attract other people to the subject. More than 100 institutions around the world are potentially interested in the topic. One question for the workshop was to assess whether this model can be exported to other parts of maths. Sage-combinat “Sage-combinat 8 is a software project whose mission is to improve the open source mathematical system Sage as an extensible toolbox for computer exploration of (algebraic) combinatorics, and foster code sharing between researchers in this area. In practice, Sage-combinat is a collection of experimental patches (i.e. extensions) on top of Sage, developed by a community of researchers. The intent is that most of those patches get eventually integrated into Sage as soon as they are mature enough, with a typical short lifecycle of a few weeks. In other words: just install Sage, and you will benefit from all the Sagecombinat development, except for the latest bleeding edge features. ” Sage-combinat has around 30 contributors. Sharing software and data in maths A fraction of the community represented in the workshop considers that Sage fulfils its needs. Others want to develop added-value services and databases, aggregating information from Sage and eventually additional information. Facilitating the usage of Sage is one goal, and the construction of a “Sage Explorer” using the semantic information in Sage to allow exploration of Sage objects and connection between them was prototyped. The workshop concluded with suggestions for future projects, one being a major conceptual evolution for LMFDB: that all the code handling mathematical calculations should be implemented in Sage, and just called by LMFDB. This could be a path for development of other added-value services in other domains. The workshop was based on a “hands-on” approach, with topics for discussions selected each day, discussions between the interested participants and rapid construction of prototypes to assess ideas and feasibility. General objectives are well understood and possible implementations are assessed in this bottom-up way, which seems to correspond to the disciplinary culture. These projects demonstrate an excellent expertise in building and managing shared software collections at community level. 6 7 8 http://www.lmfdb.org/bigpicture http://www.aimath.org/knowlepedia/ http://wiki.sagemath.org/combinat rdaeurope@rd-alliance.org | europe.rd-alliance.org 36 37 Community Data Analysis: Huma-Num Goal of analysis The goal of the analysis is to present the case of a research community relatively new to “digital science”, in a domain, humanities and social sciences, which has several ESFRI programmes in the data management and dissemination field. Huma-Num is the French national infrastructure and project in the domain, which has its own goals and methods, and which also acts in support to the future ERIC DARIAH and ESFRI CLARIN project (by the way of Aix-Marseille University). Analysis provided by CNRS. Description of the community Huma-Num (http://www.huma-num.fr) is a “Very Large Research Infrastructure” (Très Grande Infrastructure de Recherche or TGIR) labelled by the French Ministry of Higher Education and Research. It was created in March 2013 by the merging of two TGIRs in the domain of social and human sciences, ADONIS (2007) and CORPUS (2011). Its aim is to facilitate the switch to numerical science for the social and human science communities. Human-Num is managed by the CNRS in association with Aix-Marseille University and Campus Condorcet, and the target community is the research and teaching community from CNRS, plus University teams, in the domain. The activities include (1) creation of and support to communities organised thematically (“consortia”) for their adaptation to data conservation and dissemination, and dissemination of technologies and methods so that they can become actors (data and tool providers) in the process; (2) provision of massive storage and long term archiving, and of data dissemination and browsing capacities. Context The humanities and social science community is a newcomer in the domain. It produces very significant data volumes which obliges it to re-think its methods and to implement a new kind of data management with a data life cycle including re-use and medium/long term conservation. Research across the sub-discipline borders is also an incentive. Data types Data types addressed by Huma-Num are very diverse, and include modern and ancient texts, fixed and animated images, audio and movie pictures data, very large data series from surveys, 3D data obtained from in situ numerical captors or reconstructed, etc. Data formats One aim is to unify data formats using widely used formats, e.g. XML/TEI for texts, MPEG4, MATROSKA for motion picture, XML/EAD and XML/METS as envelope of complex data, etc. Data are annotated (in XML/TEI format for texts for example) by the data producers or by the researchers which edit text or image corpuses. rdaeurope@rd-alliance.org | europe.rd-alliance.org 37 38 Data life cycle The data life cycle follows the OAIS model for the long term preservation section. Data organization Research produces corpuses, which are documents organised into a set which has a scientific meaning. A corpus can be a researcher’s (including its archival materials), or from a laboratory, field campaign or science & culture heritage project, a survey, etc. Dissemination can be through a platform specific to a community or a discipline, or through the ISIDORE platform developed by Human-Num, which provides a global unified access to data with harvesting, enrichment, links between data, data browsing and APIs and SPARQL endpoint. Data exchange, data services and tools Data Exchange is through APIs provided by the specific community or through the general ISIDORE service. One aim is to expose in the linked open data (with RDF) data and metadata description with scientific communities ontologies. For this, ISIDORE service proposes an assembly line to annotate, turn in RDF, enrich with linked data URIs (dbpedia, Geonames, VIAF, etc.) and expose results in an SPARQL endpoint (http://www.rechercheisidore.fr/sparql). Legal and policy issues Huma-Num advocates open access, including for software. When communities enrich raw data the recommended licence is the Etalab Licence Ouverte/Open Licence (http://www.etalab.gouv.fr/pages/Licence_ouverte_Open_licence-5899923.html), which is compatible with the Creative Commons licences. Challenges Among the challenges is the massive scale of the task since the aim is a change in paradigm in the way this very diverse community deals with its data productions. One important goal is that long term data aspects are systematically taken into account before data production begins. Human-num is working at increasing community awareness but then time and efforts will be needed before adoption by the whole scientific community in the humanities and social sciences. The adoption level currently is very uneven between the different sub-disciplines. Everywhere there are individuals which care about data, but it is not the case for all subdisciplines as communities. One can hope in a kind of “snowball effect” with the most advanced disciplines and individuals progressively motivating their neighbours. Community Data Analysis: INAF centre for Astronomical Archives Goals of analysis The goal is to analyze activities of a medium sized astronomical observatory, Trieste Observatory, which has responsibilities relevant to scientific data at the national and international level. Analysis provided by CNRS. Description of the community rdaeurope@rd-alliance.org | europe.rd-alliance.org 38 39 The Osservatorio Astronomico di Trieste (OAT) performs data management tasks in addition to research in astronomy. It hosts the INAF centre for Astronomical Archives (IA2). IA2 manages Italian data from ground based telescopes, in particular the Telescopio Nazionale Galileo and the Large Binocular Telescope. The Telescopio Nazionale Galileo is an Italian Telescope based in the Canary Islands, which hosts among its instrument an international one, HARPS-N (High Accuracy Radial Velocity Planet Searcher). The Large Binocular Telescope, based in Arizona, is an international collaboration gathering agencies and laboratories from Italy, USA and Germany. IA2 also hosts data from research teams and individuals. OAT hosts VObs.it, the Italian Virtual Observatory project. Goals of the community Host data, provide data to the astronomical community and work on interoperability standards and tools. Context: Current practices - achievements and limitations: IA2 hosts raw data provided by the telescopes and some reduced data. They are in charge of the data pipeline which produces reduced data of the HARPS-N instrument. They keep the data archives and distribute public data. Each telescope defines its data policy. The data is in general made public after one year, and metadata are immediately public in all cases. IA2 also hosts private data from research teams and individuals. Data types Images, spectra, radio data, catalogues. FITS (the reference format for astronomical data) is the standard. Data organization and exchange Data is accessible through web interfaces and through the IVOA protocols. All public data is made accessible in the astronomical Virtual Observatory. Data services and Tools Public data is available through the VO-enabled tools and through web interfaces. IA2 has developed the VODanse system to build VO-enabled databases. The system is used internally and there are plans to release it. Data ingestion is through the home-made NADIR data handling system which allows management and ingestion with large data rate over several geographic sites. The ingestion part of NADIR is based on the OAIS standard. Legal and Policy Issues The Italian policy is that data from a telescope is made publicly available one year after observation. The private period can be expanded on demand for programs which last for a long time. Limitations /issues There are no difficulties with telescopes since a MoU defining their relations with IA2 is signed. In some cases data provided by teams or individuals are not well formatted. A minimum standard (FITS data files and VO keywords) is required. rdaeurope@rd-alliance.org | europe.rd-alliance.org 39 40 Community Data Analysis: ML-group Data-Management in an AI group Example: ML-group, Inteligent systems, icis, RU (Tom Heskens, Josef Urban) Analysis provided by MPI-PL General Data Flow The ML groups does not collect any data on their own. They work together with other groups that do collect data such as the Donders institute, bioinformatics at the university itself and many others. Typically very little is done to the data. After receiving the data it is cleaned up and stored on disk. Experiments are done to the data on disk. In general nothing further is done to the data and no effort at long-term preservation is made. The only computation that is done on the data (outside of the experiments) is cleanup; it will be discussed below. In general the group deals with the technology side of the research, not the data side. Computational Steps The data as it comes in generally needs to be cleaned up. The extent of the data cleanup depends on the type of data and the quality. Typically unit conversion (e.g. degrees Fahrenheit to Celcius), and removing clearly erroneous measurements is done. Some data is already partially cleaned up, MRI data often is. Example Projects The exception to the rule is the project on mathematical proofs led by Josef Urban. The project is developing a system to aid in the creation of mathematical proofs. That is, given a (hard) mathematical theorem the system tries to find a proof or an inconsistency (given as a counterexample). Computationally this is extremely difficult. The novel approach of the system is that it is non-exhaustive and it tells the user when it is too weak to solve the proof/disproof. The uniqueness of the system lies in three aspects: 1) it does not do complete calculus, 2) It tries to learn techniques from pre-existing proofs found in the libraries, and 3) it limits the search-space. This system needs data, as input it takes three pre-existing datasets and uses them as a base. This comes with two major problems: translation and inconsistencies. First, these mathematical datasets (attempts to encode all mathematical knowledge in a computerreadable way), are written in different formalisms, such as type theory, high order logic, or set theory. In the project this is all stored in first-order-logic. Second, these datasets are not wholly consistent between each other or even internally. Each dataset has a core (kernel) part, which is consistent. The inconsistencies mainly arise in more complex theorems and definitions. The use of such proof-systems lies in their ability to formally look at the correctness of a given theorem given the base knowledge. This has many applications, from chip design to the verification of software systems. rdaeurope@rd-alliance.org | europe.rd-alliance.org 40 41 Data Reuse Data is reused as much as possible, also in between projects as to maximize the number of publications. Community Data Analysis: Computational Linguistics Group Example: CL group of Antal van den Bosch @RU Analysis provided by MPI-PL General Data Flow The CL group, part of the Language and Speech Technology PI group of the Centre for Language Studies (Faculty of Arts, Radboud University) bases most of its research on textual corpora, experimental data from others, web crawls (e.g. Twitter, Wikipedia, etc.). The data is lightly pre-processed: Usually only tokenization and conversion to a standardized formal (Folia9) is done. The LDC 10is a major source for the corpora. The group has a corpora store, in which the processed data should end up. This is an informal and not enforced policy. In practice most corpora end up on it relatively quickly. Most data crawled from the internet is easily stripped from HTML and so forth. Data from corpora and others comes in a variety of data formats such as TIE, IMDI, CMDI, Alpino, Plaintext, etc. and will have to be converted. A library of converters is maintained for this purpose. FoLiA is an XML-based annotation format, suitable for the representation of linguistically annotated language resources. See also: http://proycon.github.io/folia/. 10 The Linguistic Data Consortium creates and distributes a large number of language resources. 9 rdaeurope@rd-alliance.org | europe.rd-alliance.org 41 42 Crawler Corpus Tokenize + Folia Store Experimental Data Others Experiment Computational Steps In collaboration with the CL group at Tilburg University, the group maintains a Dutch language computational suite for standard computational linguistic analysis steps called Frog. For their own processing this suite is most frequently used for Tokenization and packaging (into the Folia format). As needed for experiments this suite is used to add more annotations as well. In general machine-learning-based modules that have been trained in advance on annotated corpora and lexica perform the computational steps. A master process combines all module outputs and casts them into FoLiA. Typically the word forms are used in a sliding window 11. Example Projects In Adnext (part of the COMMIT programme) the group works together with a major news service (ANP) to recognize patterns in text (ANP and Twitter) in order to predict events. Data from the ANP news service is combined with data from Twitter to see what people write around events. Events are defined by the fact that the news service wrote about them. Harvested patterns are used to try and predict events from current Twitter data. Data Management As mentioned above processed data is placed in a central store to have a “fixed” version to refer to and use. This store consists of a file system and is managed by hand. In the sliding window approach the focus slides over all available words oneby-one, for each focus word also taking into account some immediate context words. 11 rdaeurope@rd-alliance.org | europe.rd-alliance.org 42 43 Provenance information of data is managed in an ad-hoc manner. I.e. every researcher in the group has his or her own way to handle provenance. This ranges from keeping detailed logs of all operations and experiments to less structured methods. Data Reuse A large amount of data reuse occurs and is encouraged. In CL Data reuse makes experimental results comparable between systems. I.e., if two systems for the same task are tested on the same datasets you can directly compare the results from the publications. Failing that one has to run one’s own tests. The group find that publishing their data, results and programs on the web (e.g. Github for codebases) results in more cooperation, more citations, and also use. Community Data Analysis: CLST CLST group at RU General Data Flow CLST (Centre for Language and Speech Technology) deals with both text and audio data. Partially they make their own recordings; partially these are done via web applications, such as in the case of Wikispeech. In general data is stored at the university and often it is published as a corpus via LDC. Computational Steps The computational steps taken depend on the data and the goal that is had for the data. In general a non-computational step done on most data is that of transcription. Sometimes transcription can be done automatically, for example when prompt files (for teleprompters) are available. Phonetic transcription can sometimes be done automatically, depending on the type and the quality of the audio. Metadata is added as a computational step and also edited by hand. The resulting data, that is transcriptions and metadata, are used to build combinations databases. Concurrently with the databases the audio recordings are stored as files. These databases are then used to process queries on the data and to select specific intervals of audio for further analysis. Example Project An example project for the CLST is the Oral History Annotation Tool. With this tool it is possible to search and annotate historical audio dealing with the second world war, Dutch Indies, and New Guinea. These data is curated at DANS. Data Management There is a corpora disk for internal use. Finished projects are managed externally in the form of corpora. Usually at the LDC, TST or at the project partner. All data is made public, via corpora or via the web. rdaeurope@rd-alliance.org | europe.rd-alliance.org 43 44 Internally the data is managed by the system administration. That means, there is a backup, but nothing specific for the type of data. Data Reuse Reuse is extremely common, but dependent on the dataset, for instance CGN (Corpus Spoken Dutch) is reused extremely often. As mentioned above the data is also made available publically. This is advertised on the CLARIN infrastructure as well. Community Data Analysis: Donders Institute Analysis provided by MPI-PL General Data Flow A large amount of the data in the institute is self-generated for a specific research question. This data comes from several sources, most notably: MRI, MEG, EEG, NIRS, behavioural studies, and physiological (eye-tracking, ECG, EMG). The generation and processing of the data happens in a linear process. After planning an experiment, measurements are taken. The data is preprocessed and then analyzed. The pre-processing is ad-hoc and differs based on needs. The measurements, and results are archived. Archival is file-based and there is no metadata. A project typically has 20 to 50 subjects. More and more longitudinal studies are performed. Computational Steps Typical preprocessing steps include motion correction, noise filtering, and compression on a per-subject basis. Over the experiment or a collection of subjects statistical analysis is performed. Example Project A large meta-project happening within the Donders institute is the BIG (Brain Imaging Genetics) project, which aims to look at the relation between genetics and the brain. Currently the project has some 2500 brain scans and some 1300 gene sequences. For statistical analysis from brain structure to genetics more than 10.000 pairs of scans and sequences are needed. That is, to generate hypotheses’. However, even with the limited data currently available, it is already possible to work based on hypothesis. Usually they postulate relations between the presence or absence of certain genes and volumes or connectivity of brain areas. This has already publications.html . resulted in many publications: see http://cognomics.nl/big/big- rdaeurope@rd-alliance.org | europe.rd-alliance.org 44 45 Data Management Data is stored centrally and backup up daily. Next to the central data all data is archived on portable harddrives. All mutations to the data are cataloged by hand. Data Reuse Data reuse within the group is common. External reuse is uncommon due to privacy laws and ethics. These days the consent forms signed by the subjects are broader, enabling more cases of reuse. There are some external cooperation with other institutes, in such cases an MOA is present. All communication on existence of data happens within the scientific dialogue, there is no search system. Community Data Analysis: Meertens Institute Analysis provided by MPI-PL General Data Flow In dataflow depends on the type of data. In the figure we show the normal dataflow for scanned and OCRed (historical) text. Next to texts other types of data are frequently added such as dictionaries. Typically the cleanup of OCRed text is done automatically with the TiCCL tool. Afterwards, if applicable, further automatic and manual annotations are added such as lemmata, part-ofspeech tags, named entities, and increasingly opinion mining. Naturally detailed metadata is added. This whole: primary data, secondary data (annotations), and metadata is stored. Next to this set the possibility exists these days for users to generate their own annotations and store these in the system. These are stored separately. The Meertens institute has developed and uses profiles to ingest data into the system. That is, if they have ingested the data-type before they have a largely automated workflow for ingesting it. rdaeurope@rd-alliance.org | europe.rd-alliance.org 45 46 Scanning Images Main Store User Annotation OCR Cleanup User Store Annotation Figure 1: Typical DataFlow in the Meertens institute. Note that it is possible for users to have their own, extra, layers of annotation. Computational Steps Typical computational steps taken are computational linguistic processes. Frequently used are: Tokenization Automatic clean-up of text (using hashing collisions) Lemmatization Part-of-Speech tagging Sentiment mining Named entity recognition Metadata Generation Example Projects Nederlab is a project that aims to make available all digitized dutch texts to researchers and students. The collection ranges from 800 to the present. Within Nederlab the fully automated process is combined with human curators. These curators control the data quality and make sure that texts are linked to the right author-entity for instance. Within Nederlab researchers will be able to construct their digital workflows in order to postprocess the selected data. The resulting processes can be stored for further use. The processes can result in further annotations on the primary data. These annotations can also be stored for further use. Data Reuse All data in the system is inherently reused as the Meertens deals with historical texts. Some specific steps are taken to encourage reuse and interoperability such as the tagging of datacategories with Isocat categories, the creation of an institute-names lexicon (using openskos). rdaeurope@rd-alliance.org | europe.rd-alliance.org 46 47 Making sure the data is findable encourages reuse. One of the ways the findability of the data is increased is by the embedding of the Meertens institute in the European CLARIN infrastructure. Community Data Analysis: Microbiology Example: Molecular Biology department of MPI for developmental biology (Jonas Müller, Joffrey Fitz, Xi Wang) Analysis provided by MPI-PL General Data Flow Data at the Molecular Biology department comes in three types, namely: 1) sequencing data (DNA and RNA), 2) automated images of growing plants, and 3) small scale imagine, e.g. of guppies, microscopy-photography, plants in environment. Of these the first generates by far the largest amount of data. For the sequencing data systems are in place for data management upon release, the other types are generally released as online supplements to publications without any specific data management. In the rest of this short report we will focus on sequencing data. Sequencing data is produced in a raw form on the sequencing machine. On the machine itself it is converted to a usable format and that data is copied to a central storage. Each project has an pre-allocated amount of storage. The storage of each project is associated with a detailed description of the project, though there is not fixed standard for this. If the data is no longer needed for active research it is moved to a tape archive. In case of RNA sequences the environment of the sample is pertinent to the sequences. Typically time series with controlled environment are used. Computational Steps Sequencing data is filtered after it is copied from the sequencer. The sequencer attaches a quality value (probability that the base is correct), the bases are discarded if the probability value is too low. Typically the short sequences that come out of the machine are also aligned, this happens in two ways: 1) by comparing the sequences to a known reference sequence to determine the absolute position on the genome, or 2) by Denovo assembly, which typically results in an incomplete genome with many possible parts of sequences, this is still usefull. For the main subject of the research of the microbiology group (Arabidopsis thaliana), there is a reference sequence with many annotations on the functions of genes and many other aspects available. The sequences found in the sequencing step are addressed using the absolute positions they take on the refence genome. Interesting for research are the differences between the sequences genome and the reference genome. These variances can take the form of deletions, inversions, or single base differences. rdaeurope@rd-alliance.org | europe.rd-alliance.org 47 48 Example Projects One project spanning five years looked at the rate of mutation in Arabidopsis Thaliana for 30 generations 12 . Many sequences where needed for this project as well as good data management. This found that “Our results imply a spontaneous mutation rate of 7 × 10−9 base substitutions per site per generation, the majority of which are G:C→A:T transitions. We explain this very biased spectrum of base substitution mutations as a result of two main processes: deamination of methylated cytosines and ultraviolet light–induced mutagenesis”. Another example project is ongoing. For this some 80 worldwide Arabidopsis Thaliana where sequenced. The aim of this project is to cross all 80 varieties to study viability. This will help isolate genetic factors which make certain crosses unviable. One important factor is the amount and type of immune-system genes in the cross. Data Management Data management internally is done in two stages. First, it is stored on the working filesystem, with backups while the data is being used. Second, once an article is published about it, the data is made available over the internet. Typically the data is made available using SRA or ENA. All published data is available freely over the internet. For microbiological genetic data a PID is assigned from a new PID system that is being currently developed by the community. Data Reuse Reuse of raw data is rare for two reasons: 1) the subjects of the research (Arabidopsis Thaliana) are very easy to grow, and 2) the genetic sequencers get much better over time. The result is that if similar data is needed it is usually better to redo the relevant parts of the experiment. The resulting data is better which makes analysis much easier. On the other hand, analysis results of the data are reused frequently. These are usually available as a tabseparated-file. Community Data Analysis: Arctic Data Example: NIOZ Analysis provided by MPI-PL General Data Flow Their ship the Pelagia collects most data that arrives at the NIOZ institute. During a voyage two things are collected: 1) measurements, and 2) samples. Measurements are varied, but the typical one is CTD: Conductivity, temperature, and depth. The samples consist of water and soil samples for which location, depth and so on are recorded. The measurements are preprocessed on return of the ship at the institute and then stored in the relevant databases and stores. 12 DOI:10.1126/science.1180677 rdaeurope@rd-alliance.org | europe.rd-alliance.org 48 49 The samples are stored in the institute on return of the ship. The sample locations, depth, age, and type of storage are stored in the archive. If the sample is analyzed at some point the output of that analysis is stored in the archive. Furthermore, in the field, data is made available real-time to third parties. This is for instance used for the calibration of Argo systems. The Argo system consists of autonomous underwater robots that surface once every 10 or so days and that have to calibrate their instruments at that point. All the collected data is made available on the Internet. There are databases and stores of several types including: CTD data, anchor-points, sediment data, optical measurements. Available via www.nodc.nl. Samples Storage Measurements Measurements Preprocess Archive Pelagia publish Figure 2: Overview of the rough dataflow in the NIOZ institute. The "archive" consists of a number of databases and stores. Preprocessing is done in many ways by different departments. The dashed line indicates that this step happens at an undetermined time. Computational Steps The main computational steps are taken during the pre-processing of the CTD data. The raw measurements output by the instrument are converted to units and cleaned up. Cleaning up consists of, amongst other steps, calibration to known-good values and the removal of spikes. After cleanup the metadata is checked and added. Furthermore metadata on “meetstations” (measuring stations) is added. A “meetstation” is a place on the voyage where the ship has maintained a fixed position for some time in order to do measurements. Specific computational steps for specific types of data (e.g. sediment analysis) are performed by the relevant departments within the institute. rdaeurope@rd-alliance.org | europe.rd-alliance.org 49 50 Example Projects A new project is currently underway to integrate the efforts of the NIOZ wrt the Waddenzee with the efforts of other entities wrt the Waddenzee. Other major partners in this project are the universities of Nijmegen and Groningen, IMARES, and SOVON. Their goal is to join up their project with ILTER and LTER-Europe. The project will make all data collected in the Waddenzee, both ecological and socioeconomical available at one central web-location. Data Management Data is stored in two separate locations in the institute. These locations are at the extreme ends of the building. The usual steps of multiple stores and backups are used. Due to the size of the building, it is extremely unlikely to lose both data centers. Data Reuse The collected data as made available on the website is reused for research both inside the institute and outside the institute. Furthermore the samples that are stored at the institute are also reused for different projects. After all, it is much cheaper to analyze pre-existing samples compared to going out to the arctic to take new samples. The possibility for data-reuse is a major source of new cooperative projects for the institute. The publication of their data results in researchers approaching NIOZ for collaboration. This is a clear benefit. rdaeurope@rd-alliance.org | europe.rd-alliance.org 50