Experiment 4: Colorimetric Determination of Protein Concentration

CHEM 240 L Lab 2 Spring 2011
Lab 2: Colorimetric Determination of Protein Concentration
Lab Objectives
To determine the concentration of protein in an unknown sample
To become familiar with the Beer’s Law and use of a spectrophotometer for analytical experiments.
To understand the concept of a colorimetric assay and its limitations.
To understand the concept of a standard curve and how to assess its quality.
Be able to determine whether to omit a data point using Grubbs’ Test and experimental observations.
Be able to use LINEST and STDEV functions in Excel.
Reading from Exploring Chemical Analysis. Harris, D.C. 4 th ed.
Ch. 18-2, pp. 396-400: Beer’s Law.
Ch. 18-3, pp. 402-404: practical considerations when using a spectrophotometer.
Ch. 3-6, pp.77-78: plotting data in Excel.
Ch. 4-1, pp. 84-66: a basic introduction to standard deviation.
Ch. 4-4, pp. 92-93: Grubbs’ Test.
Ch. 4-6, pp. 96-98: for constructing and using a calibration curve.
Ch. 4-7, pp. 98-100: how to set up a spreadsheet for Least Squares analysis
Pre-Lab
1) Why is incubation at 37
C for 30 min. important to this assay?
2) Describe three possible sources of error that you might encounter in this lab and explain how you could mitigate them.
3) Generate a table in your notebook similar to the table in Part B Step 2. Decide which
BSA stock solution to use for each standard and fill in the volumes of the BSA stock solutions and water.
Introduction
There are a number of common methods for the determination of protein concentration.
The simplest relies on the absorbance of the protein at a wavelength of 280 nm, but this depends on the presence of aromatic amino acid side chains, primarily tyrosine and tryptophan, which are not always in high abundance in proteins. Furthermore, the extinction coefficient,
, of a particular protein is sensitive to buffer conditions (pH, ionic strength, detergents, etc.). More sensitive and universally applicable concentration determination assays rely on binding of dyes directly to the protein (the Bradford method) or the reduction of copper (II) ions by protein and the subsequent binding of a dye to the reduced copper (I) ions (Lowry method, Biuret method, bicinchoninic acid (BCA) assay). Each of these assays relies on a similar general principle: when the dye in use binds either to protein or to the reduced copper ions, it undergoes a color change. The number of dye molecules undergoing the color change is linearly proportional to the protein concentration over a range of protein concentrations.
Each colorimetric assay has its limitations and advantages. The Bradford assay uses the common Coomassie Brilliant Blue dye, which makes it convenient. However, it is sensitive to the amino acid composition of the protein and a plot of absorbance vs. concentration using it is only linear over a short range of protein concentrations. The BCA assay and other copper
1
reduction assays are powerful, linear over a wider range of protein concentrations and are far less sensitive to the identities of amino acids in the sample, but take longer to do and require more uncommon chemicals. The bicinchoninic acid (BCA) assay is generally considered to be the easiest of the copper reduction assays, and is the method we will use in today’s experiment.
Regardless of the method used, colorimetric assays require a calibration process prior to determination of an unknown protein concentration. This process requires the construction of a standard curve. In this process, the assay is run on several known concentrations of a standard protein, often bovine serum albumin (BSA) or bovine gamma globulin (BGG), and the absorbance values for the known concentrations are measured. Ideally, plotting absorbance vs. concentration will give a linear plot. The assay is then conducted on a protein sample of unknown concentration, the absorbance is measured, and, assuming the absorbance value is somewhere within the linear range of the standard curve, the value for concentration that would correspond to that absorbance is determined. Assuming minimal systematic error, the better
(more linear) the standard curve, the more accurate the concentration determination.
The BCA assay (Figure 1) relies on the temperature-dependent reduction of Cu
2+
ions by peptide bonds in the protein in an alkaline environment. The side chains of the amino acids cysteine, tyrosine, and tryptophan also participate in the reduction of Cu
2+
under alkaline conditions. The organic dye bicinchoninic acid is then added, which selectively chelates Cu
+ ions, leading to the formation of a metal complex with a strong purple color. This Cu
+
-BCA complex absorbs light at 562 nm and can be measured with a UV-Vis spectrophotometer.
In this lab, you will test the operating protein concentration range of the BCA assay by generating a standard curve using known concentrations of BSA. You will assess your data for its linearity, perform a linear regression, and use your trendline to determine the concentration of two unknown protein samples.
It is important to note that the color formation in the BCA assay depends on more than simply the number of peptide bonds present in the sample. It is also affected by the structure of the protein and the presence of the amino acids cysteine, cystine, tryptophan, and tyrosine. If the unknown protein sample has similar characteristics to the protein measured in the standard curve, the concentration of the unknown can be determined with a high degree of accuracy. However, if the unknown protein sample has many different characteristics to the protein measured in the standard curve (in size, amino acid composition, or structure), the error in the concentration determined may be significant. In this lab, the unknown protein sample is actually a sample of
BSA, so you will be comparing the same protein as in your standard curve, which is the ideal way to determine protein concentrations.
It is also important to be consistent in your experimental technique. Treat all samples equally. Pipetting should be performed as uniformly and reproducibly as possible. Chemical reactions generally proceed faster at higher temperatures. It will be particularly important for all samples to be incubated at elevated temperatures (37
C) for as close to the same time interval as is practical.
2
Step 1:
Protein + Cu 2+
OH -
Protein + Cu +
Step 2:
Cu + + 2 BCA
-OOC
N
N
Cu +
N
N
COO-
COO-
Figure 1: The BCA assay procedure. Addition of copper
(II) ions to a protein solution in an alkaline medium reduces the copper (II) ions to copper (I).
BCA added to the solution chelates copper (I) in a 2:1 stoichiometry. The BCA-Cu + complex produces a strong purple color.
-OOC
Experimental Procedure
Work in groups according to your instructor.
A. Preparation of Working Reagent
1.
You will need to make at least 11 mL of working reagent per person in the group.
2.
BCA Reagent A contains sodium carbonate, sodium bicarbonate, bicinchoninic acid, and sodium tartrate in 0.1 M sodium hydroxide. BCA Reagent B contains 4% copper (II) sulfate. Prepare the working reagent by mixing 50 parts of BCA Reagent A with 1 part
BCA Reagent B in a 50 mL disposable centrifuge tube. Be extremely careful not to cross-contaminate the Reagent A and Reagent B stocks!
3.
Mix well by inverting the capped tube several times. The initial solution will be turbid, but should make a clear green solution after mixing.
B. Preparation of Standards
1.
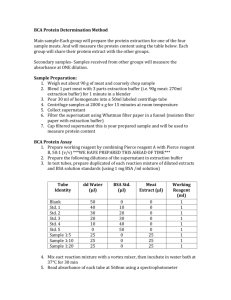
Your group will run three trials of everything, so you will have to prepare three complete sets of the above series. Therefore, each member in a group should construct their own series of standards.
2.
In two separately labeled and clean microcentrifuge tubes, obtain a sufficient amount of the 5.00 mg/mL bovine serum albumin (BSA) stock and a sufficient amount of the 1.00 mg/mL BSA stock solutions.
3.
Make nine 200
L standard solutions covering a range of BSA concentrations of 0.00 –
2.00 mg/mL. Have the following table reproduced and completed in your lab notebook prior to lab ( see Pre-Lab Question 3 ).
3
Tube Number
S0
S1
S2
S3
S4
S5
S6
BSA Stock Added (
5.00 mg/mL
L)
1.00 mg/mL
Water Added
(
L)
Final [BSA]
(
g/mL)
0
100
200
400
600
800
1000
S7
S8
1500
2000
4.
Unknowns: Obtain 200
L of U1 and 200
L of EITHER U2 or U3.
5.
For each Trial: In 1.5 mL microcentrifuge tubes, prepare a total of 11 samples as follows: Add 50
L of the appropriate sample and add 1000
L of BC-WR. Vortex gently to mix.
Below is an example of the beginning of a table in your notebook.
Tube Number Sample (
L) Sample Identity Working reagent (
L)
“Your initials”
0 50 S0 1000
“Your initials”
8 50 S8 1000
“Your initials” U1 50 BSA Unknown 1000
C. Determination of Standard Curve
1.
Incubate all samples in a 37°C water bath for 30 minutes.
2.
While your samples are incubating, initialize and calibrate the Avantes AvaFast Fiber
Optic Diode Array Spectrophotometer (refer to the separate Avantes Spectrophotometer instructions).
3.
When your samples are done incubating, remove them from the water bath, and let them cool to room temperature. a.
If you do not immediately measure the absorbance, store your incubated samples on ice.
4.
Use the same five cuvets to make all your measurements. a.
Start with your blank sample and progress through the standards from least to most concentrated. Measure the unknowns last. b.
Pipet 1 mL into the cuvet and measure the absorbance spectrum from 200-700 nm.
Assess the quality of the spectrum (baseline flatness, noise level, peak shape) and make necessary notes in your observations. c.
Record the absorbance of your samples at 562 nm.
4
d.
After each measurement, return the contents of the cuvet to the sample tube, rinse the cuvet at least 3 times with DI water, and vigorously shake dry to minimize dilution of your next sample.
D. Plotting Your Calibration Curve and Determining the BSA Unknown Concentration
(Refer to the appropriate sections in the ECA text for guidelines)
1.
For the three sets of standard curve data your group has collected (Trials 1-3), plot A
562 vs. BSA concentration (
g/mL) in a spreadsheet (one plot per trial).
2.
For your individual standard curve, have your spreadsheet plot a trendline to your data and determine the following using the LINEST function:
Slope and y-intercept of the best linear fit to the data
Equation of the resulting linear fit
R
2
value for the resulting linear fit
Error in the slope and y-intercept
Plotting a trendline is done by making the plot and then selecting the data series in the chart. From the Chart menu, select “Add Trendline”. In the resulting dialog box, make sure “linear” is highlighted. Click the Options tab and click the checkboxes for “Display equation on chart” and “Display R-squared value on chart”. Hit OK. Do not force the trendline through the origin.
R
2
as well as the standard deviation in the calculated slope are measures of how well the data fits a linear trendline. An R 2 value of 1 is perfect fit. As R 2 decreases, it indicates the data fits a linear trendline less well. Use the calculated R
2
and errors in the slope to determine whether omitting suspect points or sets of points significantly improves the quality of the calibration curve.
Consult with your TA and/or the instructor if you are having difficulty determining suspect data points. As a rule of thumb, a more accurate trendline gives a y-intercept close to 0.
3.
Using a combination of Grubbs’ Test and your observations, determine which points to keep for your final group calibration curve. a.
Take the average of the remaining group A
562
values for each BSA concentration selected and plot this data. b.
Use LINEST to determine the equation for the linear regression and the associated errors.
4.
Use this final calibration trendline to determine the protein concentration of your unknowns. Report these values with appropriate significant figures, error, and units.
In the discussion section of your notebook, be sure to include:
1.
A chemical description of the processes underlying the assay, why you performed the assay, and what you were able to determine from it.
2.
An explanation for your data analysis, including why you used the LINEST function, why you decided to discard (or not discard) some data points from your curve, and the precision and accuracy of your data. Generally speaking, how confident are you in the accuracy of your concentrations of your unknowns?
3.
An explanation for any unexpected results and suggestions for how you might improve the experiment and/or your results if you were to repeat it.
5
Lab
2 Excel Report
Use this filename: “ Gonzaga username ” CHEM 240L_” 2 digit section number ” S11 Lab_2
Your workbook should contain the following sheets:
1) Your personal calibration and unknown data. a.
A table that converts your raw data to corrected absorbance. b.
A plot of your calibration data (A
562
vs. [BSA] (mg/mL)). Do not connect the points with a line. c.
All rejected data should be colored red and a brief one-sentence explanation for this should annotate that datum. i.
To annotate a cell with a comment: (1) highlight that cell, (2) go to the
Review menu, and (3) select the Comment item. A text box should appear. You can then hide a comment by right-clicking that cell and selecting Hide Comment . d.
A second plot of your refined data e.
A LINEST analysis of your refined personal data and the resulting equation for the line and R 2 value. f.
Calculated concentration (mg BSA/mL) for your two unknowns with associated uncertainty (%e and e)
2) Your group calibration and unknown data. a.
A table that converts the group raw data to corrected average absorbance. b.
A plot of the group calibration data (A
562
vs. [BSA] (mg/mL)). Do not connect the points with a line. c.
All rejected data should be colored red and a brief one-sentence explanation for this should annotate that datum. d.
A second plot of the group refined data. e.
A LINEST analysis of the refined group data. f.
Calculated concentration (mg BSA/mL) for the two unknowns with associated uncertainty (%e and e) using the group averaged corrected absorbance and the group calibration curve.
If you have any questions or would like me to look over your work for formatting and content, feel free to stop by during office hours or by appointment.
6