mec13120-sup-0010
advertisement

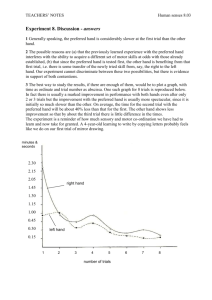
12 24 𝐿𝑖𝑘𝑒𝑙𝑖ℎ𝑜𝑜𝑑 = ∑ [( ) 0.117𝑖 (1 − 0.117)24−𝑖 ] 𝑓(𝑖, 𝑛, 𝑧) 𝑖 𝑖=0 We calculated the likelihood function starting with the probability of i number of trials scoring positive because of the non-spiked oak water. This is a binomial distribution with 24 total trials, and the probability of a single trial scoring positive because of non-spiked oak water as 0.117, which was empirically measured. As we used simulations to calculate the likelihood, we halted at i=12 trials scored positive because of the non-spiked oak water, as more positive trials should occur with probability < 10-6. 𝑓(𝑖, 𝑛, 𝑧) = 𝑝𝑟𝑜𝑏𝑎𝑏𝑖𝑙𝑖𝑡𝑦 𝑜𝑓 𝑛 24 − 𝑖 =( ) 𝑔(𝑧, 𝜆)𝑛−𝑖 (1 − 𝑔(𝑧, 𝜆))24−𝑛 𝑡𝑜𝑡𝑎𝑙 𝑝𝑜𝑠𝑖𝑡𝑖𝑣𝑒 𝑡𝑟𝑖𝑎𝑙𝑠 𝑛−𝑖 We calculated the probability of n total positive trials (as n – i positive trials caused by spiked oak water) using a binomial distribution with 24 total trials and g(z, 𝜆) as the probability of a positive trial. 10 𝜆𝑘 𝑒 −𝜆 𝑝𝑟𝑜𝑏𝑎𝑏𝑖𝑙𝑖𝑡𝑦 𝑜𝑓 𝑎 𝑝𝑜𝑠𝑖𝑡𝑖𝑣𝑒 𝑡𝑟𝑖𝑎𝑙 (1 − (1 − 𝑧)𝑘 ) 𝑔(𝑧, 𝜆) = = ∑ 𝑐𝑎𝑢𝑠𝑒𝑑 𝑏𝑦 𝑠𝑝𝑖𝑘𝑒𝑑 𝑜𝑎𝑘 𝑤𝑎𝑡𝑒𝑟 𝑘! 𝑘=0 We calculated the probability of a positive trial using a Poisson distribution to examine the number of spiked cells across wells. The average number of cells per well, 𝜆, was empirically measured for each yeast strain. z is the probability of detecting a single cell, but this probability is applied for each cell in each well. We halted at k=10 cells per well as more cells should occur with probability < 10-6. We simulated the model across values of z ranging from 0 to 2 by 0.001 increments using the n and 𝜆 values derived from each yeast strain.