mec13231-sup-0001-SuppInfo
advertisement
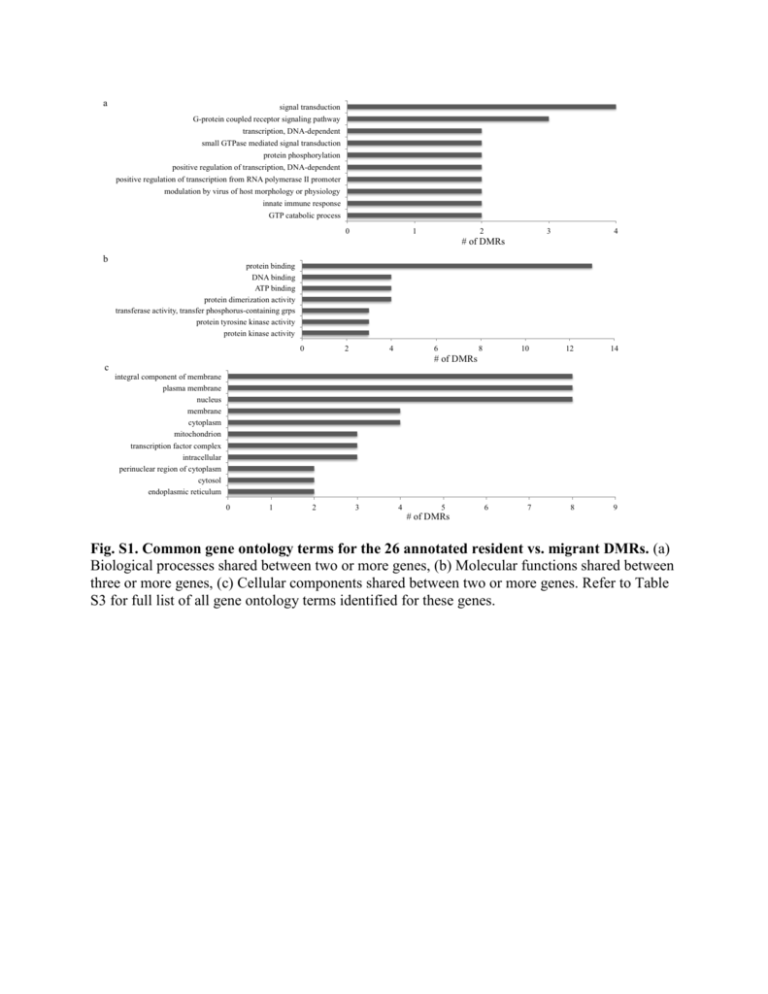
a signal transduction G-protein coupled receptor signaling pathway transcription, DNA-dependent small GTPase mediated signal transduction protein phosphorylation positive regulation of transcription, DNA-dependent positive regulation of transcription from RNA polymerase II promoter modulation by virus of host morphology or physiology innate immune response GTP catabolic process 0 1 2 3 4 # of DMRs b protein binding DNA binding ATP binding protein dimerization activity transferase activity, transfer phosphorus-containing grps protein tyrosine kinase activity protein kinase activity 0 2 4 6 8 10 12 14 7 8 9 # of DMRs c integral component of membrane plasma membrane nucleus membrane cytoplasm mitochondrion transcription factor complex intracellular perinuclear region of cytoplasm cytosol endoplasmic reticulum 0 1 2 3 4 5 6 # of DMRs Fig. S1. Common gene ontology terms for the 26 annotated resident vs. migrant DMRs. (a) Biological processes shared between two or more genes, (b) Molecular functions shared between three or more genes, (c) Cellular components shared between two or more genes. Refer to Table S3 for full list of all gene ontology terms identified for these genes. a b Fig. S2. Gene networks identified using Ingenuity Pathway Analysis. Genes color indicates methylation level for resident compared to migrant phenotype (blue = decreased, green = increased). (a) Network 1 contains 15 of the annotated DMR genes and is involved in cellular assembly and organization, cellular compromise, nervous system development and function. (b) Network 2 contains 11 of the annotated DMR genes and is involved in nervous system development and function, organ morphology, organismal development. Table S1. Read counts and alignments to French and U.S. rainbow trout genomes for resident and smolt individuals Uniquely Mapped ID # Sample Name Phenotype Sex Unfiltered Filtered (French) Sample 1 Resident_Female_2C11 Resident Female 18,867,351 16,686,974 5,670,400 Sample 2 Resident_Female_0D10 Resident Female 36,498,415 32,271,338 10,607,708 Sample 3 Resident_Male_237F Resident Male 32,286,475 28,177,042 9,502,951 Sample 4 Resident_Female_354C Resident Female 57,137,794 49,097,501 16,783,402 Sample 5 Resident_Male_3F40 Resident Male 14,493,262 12,624,913 4,202,494 Sample 6 Resident_Female_1B1D Resident Female 23,434,854 18,131,957 5,868,018 Sample 7 Resident_Male_0931 Resident Male 23,249,419 19,505,607 6,547,100 Sample 8 Resident_Male_7443 Resident Male 32,911,920 25,890,555 8,584,749 Sample 9 Resident_Male_264D Resident Male 56,913,401 45,251,857 14,664,596 Sample 10 Resident_Female_3F1D Smolt Female 86,013,558 71,046,131 24,032,778 Sample 11 Smolt_Male_3944 Smolt Male 22,280,323 19,781,458 6,617,372 Sample 12 Smolt_Female_2615 Smolt Female 16,187,577 14,407,737 4,801,539 Sample 13 Smolt_Female_0307 Smolt Female 19,471,076 17,127,341 4,969,150 Sample 14 Smolt_Male_3B14 Smolt Male 15,673,647 13,615,959 4,565,537 Sample 15 Smolt_Female_1478 Smolt Female 13,560,140 11,848,326 3,994,992 Sample 16 Smolt_Female_17D52 Smolt Female 55,188,890 42,236,943 13,782,385 Sample 17 Smolt_Female_3C59 Smolt Female 28,454,190 23,443,419 7,839,277 Sample 18 Smolt_Male_1110 Smolt Male 22,176,655 18,843,847 6,369,487 Sample 19 Smolt_Male_703A Smolt Male 4,871,811 4,326,909 1,497,464 Sample 20 Smolt_Male_6867 Smolt Male 5,989,529 5,190,837 1,774,987 No Alignments (French) 5,557,121 10,501,193 9,715,102 16,271,796 4,341,122 8,831,983 9,201,364 12,297,102 21,238,060 33,019,374 6,495,418 4,917,828 5,791,804 4,601,578 4,012,600 20,502,613 11,114,423 8,802,748 1,416,945 1,727,239 Uniquely Mapped (U.S.) 4,048,241 7,723,728 6,960,642 12,253,998 3,084,436 4,318,413 4,818,466 6,333,517 10,746,986 17,609,970 4,940,367 3,524,056 3,973,436 3,346,697 2,949,388 10,046,720 5,745,762 4,620,518 1,097,560 1,330,270 No Alignments (U.S.) 5,252,784 9,383,775 9,460,362 15,214,137 4,135,391 8,853,159 9,180,180 12,367,287 21,151,397 32,819,185 6,097,482 4,639,955 5,494,795 4,380,791 3,821,725 20,463,569 11,159,454 8,720,543 1,331,074 1,620,752 Table S2. Detailed information for differentially methylated regions (DMRs) when comparing juvenile resident and migratory O. mykiss phenotypes. DMRs with two rows of data represent hits to both French and Miller rainbow trout genomes. CpG island Start Meth diff 1 # of CpGs # of DMCs p-value q-value Contig_scaffold position Symbol Entrez gene name Functional genomic distribution Chromosome context AATK apoptosis-associated tyrosine kinase <5kb upstream of TSS Unknown open%sea -20.7 3 2 1.1E-21 1.4E-21 FR907459_3187 6454 ADHFE1 Alcohol Dehydrogenase, Iron Containing, 1 <5kb upstream of TSS Omy19 open%sea -38.2 3 2 6.2E-29 9.9E-29 MMSRT025C_2075_1 43636 ADORA3 adenosine A3 receptor Gene body Omy7 open%sea -23.5 7 4 5.4E-45 2.4E-44 FR904328_63 1466297 (23.6 7 4 3.7E(46 1.4E(45 MMSRT001B_scaff_1733_1 22090 AHR2A aryl hydrocarbon receptor 2 alpha 5-10kb upstream of TSS Unknown shore 31.9 4 3 1.1E-43 3.6E-43 MMSRT073E_1706_1 3003 CCDC93 coiled-coil domain containing 93 Gene body Omy3 shore 20.7 3 1 6.8E-18 8.6E-18 FR905754_1482 115054 CD247 CD247 molecule <5kb upstream of TSS Unknown open%sea 22.8 5 2 3.2E-34 6.3E-34 FR906353_2081 23649 CLDN10 claudin 10 Gene body Omy3 open%sea 29.5 3 3 3.0E-36 6.8E-36 MMSRT042G_1581_2 35928 CRYBB3 crystallin, beta B3 Gene body Omy7 island 21.3 4 1 1.0E-35 2.1E-35 FR904434_167 1335767 21.1 4 1 4.6E(38 1.2E(37 MMSRT117A_scaff_1889_1 19751 DDX10 DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 <1.5kb downstream of stop codon Omy10 open%sea 29.5 4 3 1.2E-31 2.2E-31 FR904727_455 284644 GEMIN2 gem (nuclear organelle) associated protein 2 Gene body Omy19 open%sea -34.1 3 2 2.5E-30 4.1E-30 FR904474_207 438741 (34.2 3 2 1.6E(30 2.6E(30 MMSRT071A_scaff_3011_1 47548 GRM1 glutamate receptor, metabotropic 1 Gene body Omy19 shore -31.3 7 5 1.2E-53 1.0E-52 FR904329_64 922718 (28.4 8 5 7.6E(48 4.1E(47 MMSRT062C_scaff_2450_1 26458 HES6 hes family bHLH transcription factor 6 <5kb upstream of TSS Unknown shore -20.7 3 1 2.2E-14 2.6E-14 FR905566_1294 184909 KIAA1467 KIAA1467 <5kb upstream of TSS Omy4 shore 23.2 4 1 1.8E-17 2.2E-17 FR905145_873 106914 23.1 4 1 1.2E(17 1.5E(17 MMSRT069B_scaff_1646_1 26732 MYOM3 myomesin 3 <5kb upstream of TSS Omy3 island -21.7 3 1 7.9E-10 8.0E-10 FR904357_92 399171 (21.7 3 1 9.0E(10 9.0E(10 MMSRT026G_scaff_1716_2 87415 NEUROD4 neurogenic differentiation 4 <1.5kb downstream of stop codon Unknown shore -31.4 3 2 8.3E-36 1.8E-35 FR906233_1961 31266 (31.7 3 2 4.3E(36 8.2E(36 MMSRT142F_scaff_3021_1 14688 NHLH2 nescient helix loop helix 2 Gene body Omy3 shore 38.0 14 12 3.2E-76 1.4E-74 FR907983_3711 31771 PALLD palladin, cytoskeletal associated protein Gene body Omy22 island 24.4 8 2 7.2E-62 7.9E-61 FR904673_401 443036 PCDHGC5 protocadherin gamma subfamily C, 5 Gene body Omy10 shore 48.8 5 5 3.1E-49 1.7E-48 FR905400_1128 190015 RASD1 RAS, dexamethasone-induced 1 5-10kb upstream of TSS Omy20 open%sea 21.5 3 1 1.8E-16 2.2E-16 FR905370_1098 150900 RGL2 ral guanine nucleotide dissociation stimulator-like 2 5-10kb upstream of TSS Unknown shore 21.2 3 1 1.2E-15 1.3E-15 MMSRT050A_1640_1 26199 RSAD2 radical S-adenosyl methionine domain containing 2 <1.5kb downstream of stop codon Unknown open%sea -61.8 10 10 2.7E-69 5.9E-68 FR908501_4229 32535 SBK1 serine/threonine-protein kinase SBK1-like Gene body Omy12 island -28.6 21 10 7.0E-86 1.9E-84 MMSRT118F_1666_1 68899 SETD3 SET domain containing 3 Gene body Omy19 open%sea 42.5 6 6 5.4E-53 4.0E-52 FR904415_148 55876 41.9 6 6 2.5E(53 2.2E(52 MMSRT017H_scaff_1643_3 51803 STYK1 serine/threonine/tyrosine kinase 1 <1.5kb downstream of stop codon Unknown open%sea 28.3 4 3 3.5E-40 1.3E-39 FR904869_597 536451 TMEM150A transmembrane protein 150A Gene body Omy16 open%sea -23.6 3 1 6.4E-24 8.7E-24 MMSRT054E_1746_6 43876 TUBA1A tubulin, alpha 1a Gene body Unknown shelf -26.9 3 3 9.1E-34 1.7E-33 FR906595_2323 32581 Uncharacterized protein Gene body Omy10 open%sea 42.0 3 2 1.2E-30 2.1E-30 FR904314_49 1898070 Uncharacterized protein Gene body Omy10 open%sea -21.1 3 1 1.3E-10 1.3E-10 FR905562_1290 143025 Uncharacterized protein 5-10kb upstream of TSS Unknown shore -21.7 3 1 2.7E-11 3.0E-11 FR905963_1691 136118 Uncharacterized protein <5kb upstream of TSS Unknown open%sea -31.7 3 3 8.3E-36 1.8E-35 FR907215_2943 53300 Uncharacterized protein <5kb upstream of TSS Unknown shore -32.3 3 2 1.4E-22 1.7E-22 MMSRT031A_2343_2 1255 Unknown Intergenic Omy16 shelf -21.0 4 1 7.9E-29 1.2E-28 MMSRT032D_1742_1 27068 Unknown Intergenic Omy23 island -33.8 4 3 1.2E-43 3.6E-43 MMSRT040D_1677_1 1103 Unknown Intergenic Omy10 shelf 20.6 3 1 1.1E-09 1.1E-09 FR904314_49 242508 Unknown Intergenic Omy20 shelf 23.7 3 1 2.0E-28 3.2E-28 FR904322_57 1755512 Unknown Intergenic Omy19 open%sea 26.2 5 4 2.1E-38 7.1E-38 FR904329_64 1826731 Unknown Intergenic Omy19 island -30.0 4 4 2.2E-42 8.9E-42 FR904351_86 1584879 Unknown Intergenic Omy19 open%sea 31.9 3 3 8.3E-36 1.8E-35 FR904415_148 592107 31.8 3 3 4.3E(36 8.2E(36 MMSRT060F_scaff_1688_1 62484 Unknown Intergenic Omy19 open%sea 29.1 5 4 5.3E-46 2.6E-45 FR904429_162 575928 29.0 5 4 1.7E(46 7.7E(46 MMSRT063E_scaff_1784_1 8960 Unknown Intergenic Omy16 shelf 21.3 3 1 3.9E-23 5.5E-23 FR904526_258 841950 Unknown Intergenic Omy29 open%sea -20.2 5 1 1.4E-31 2.4E-31 FR904607_337 911205 Unknown Intergenic Omy10 open%sea -22.3 5 1 6.7E-37 2.0E-36 FR904613_343 800481 End position 6492 43686 1466486 22279 3134 115078 23792 36051 1335800 19784 284771 438786 47593 922934 26854 184937 106945 26763 399220 87464 31311 14733 32052 443147 190108 151030 26248 32903 69369 55921 51848 536500 43999 32732 1898089 143149 136449 53349 1415 27409 1149 242747 1755633 1826844 1584920 592154 62531 576081 9113 842094 911311 800527 Table S2 Cont. - Unknown Intergenic Unknown island -23.7 (27.2 Unknown Intergenic Omy16 shelf -24.0 Unknown Intergenic Omy10 island -20.5 (20.5 Unknown Intergenic Unknown open%sea 30.5 Unknown Intergenic Unknown open%sea 20.6 20.2 Unknown Intergenic Unknown island -21.6 Unknown Intergenic Omy3 open%sea 24.4 Unknown Intergenic Unknown open%sea 26.6 Unknown Intergenic Unknown shore -31.3 Unknown Intergenic Unknown open%sea -31.0 Unknown Intergenic Omy20 open%sea 20.1 Unknown Intergenic Unknown shelf -23.1 Unknown Intergenic Omy4 island 35.4 Unknown Intergenic Omy15 open%sea 22.0 Unknown Intergenic Omy19 open%sea -26.3 1 Percent methylation differences averaged from all CpG sites within the defined region. The comparison is residents compared to migrants (i.e., smolts). 5 5 4 3 3 3 3 3 3 3 3 9 3 8 4 7 3 4 2 5 2 1 1 1 1 1 2 2 1 5 3 3 3 5 2 1 2.8E-49 3.7E(50 6.8E-38 1.9E-11 4.2E(11 5.3E-23 4.6E-11 6.8E(11 2.1E-23 8.3E-36 6.1E-28 1.5E-65 8.3E-36 6.9E-37 6.1E-33 8.7E-60 8.7E-27 7.5E-22 1.7E-48 2.5E(49 2.1E-37 2.1E-11 4.6E(11 7.3E-23 4.9E-11 7.1E(11 3.0E-23 1.8E-35 9.3E-28 2.2E-64 1.8E-35 1.7E-36 1.1E-32 1.2E-58 1.2E-26 1.0E-21 FR904682_410 MMSRT007A_scaff_1463_1 FR904705_433 FR904748_476 MMSRT057A_scaff_1596_2 FR904859_587 FR905135_863 MMSRT023H_scaff_1491_1 FR905506_1234 FR905521_1249 FR905721_1449 FR906155_1883 FR906735_2463 MMSRT010E_1934_5 MMSRT027B_2013_1 MMSRT094H_2242_1 MMSRT108D_1238_1 FR904635 680456 76264 776886 605820 35196 347754 384237 88585 241073 262928 216024 22526 26692 90125 12107 35302 74665 340968 680564 76412 776928 605846 35222 347772 384277 88625 241089 262969 216071 22653 26738 90269 12259 35391 74674 341014 Table S3. Gene ontology full details for each DMR annotated gene when comparing resident and migrant O. mykiss juvenile phenotypes Biological Process Evidence1 PubMed Reference protein serine/threonine kinase activity IEA -- GO:0005524 ATP binding IEA -- AATK GO:0004672 protein kinase activity -- -- AATK GO:0004713 protein tyrosine kinase activity AATK GO:0005515 AATK GO:0016772 protein binding transferase activity, transferring phosphoruscontaining groups ADHFE1 GO:0006103 2-oxoglutarate metabolic process ADHFE1 GO:0015993 ADHFE1 GO:0044237 ADHFE1 GO:0055114 oxidation-reduction process ADHFE1 GO:0044281 small molecule metabolic process ADORA3 GO:0001973 adenosine receptor signaling pathway ADORA3 GO:0002553 histamine secretion by mast cell ADORA3 GO:0002687 positive regulation of leukocyte migration IEA -- ADORA3 GO:0006954 inflammatory response TAS 9164961 ADORA3 GO:0007190 activation of adenylate cyclase activity TAS 9380026 ADORA3 GO:0007186 -- -- ADORA3 GO:0050850 G-protein coupled receptor signaling pathway positive regulation of calcium-mediated signaling IEA -- ADORA3 GO:0050729 positive regulation of inflammatory response IEA -- ADORA3 GO:0043306 positive regulation of mast cell degranulation IEA -- ADORA3 GO:0070257 IEA -- ADORA3 GO:0014068 positive regulation of mucus secretion positive regulation of phosphatidylinositol 3kinase signaling IEA -- ADORA3 GO:0008016 regulation of heart contraction TAS 9837869 ADORA3 GO:0009611 response to wounding TAS 9837869 ADORA3 GO:0007165 signal transduction TAS 8399349 AHR GO:0001568 NAS 19538249 AHR GO:0000122 blood vessel development negative regulation of transcription from RNA polymerase II promoter IEA -- AHR GO:0006355 IDA 10395741 AHR GO:0006357 regulation of transcription, DNA-dependent regulation of transcription from RNA polymerase II promoter IDA 15681594 AHR GO:0006366 transcription from RNA polymerase II promoter IDA 10395741 AHR GO:0006915 TAS 12213388 AHR GO:0045893 apoptotic process positive regulation of transcription, DNAdependent -- -- AHR GO:0007049 cell cycle TAS 12213388 AHR GO:0048732 gland development -- -- Gene Symbol ID GO term AATK GO:0004674 AATK IEA -- IPI 14521924 -- -- TAS -- molecular hydrogen transport IDA 16435184 cellular metabolic process TAS -- -- -- TAS -- -- -- IEA -- Table S3 Cont. AHR GO:0030522 intracellular receptor signaling pathway IDA 10395741 AHR GO:0045899 positive regulation of RNA polymerase II transcriptional preinitiation complex assembly IEA -- AHR GO:0030850 prostate gland development IEA -- AHR GO:0030888 regulation of B cell proliferation IDA 15681594 AHR GO:0010468 regulation of gene expression IDA 15681594 AHR GO:0048608 reproductive structure development -- -- AHR GO:0006950 response to stress IDA 7961644 AHR GO:0009410 response to xenobiotic stimulus IDA 7961644 AHR GO:0007165 signal transduction -- -- AHR GO:0006805 xenobiotic metabolic process TAS 19538249 CD247 GO:0007166 cell surface receptor signaling pathway -- -- CD247 GO:0016032 TAS -- CD247 GO:0019048 viral process modulation by virus of host morphology or physiology IEA -- CD247 GO:0031295 TAS -- CD247 GO:0038096 T cell costimulation Fc-gamma receptor signaling pathway involved in phagocytosis TAS -- CD247 GO:0045087 innate immune response TAS -- CD247 GO:0050690 regulation of defense response to virus by virus TAS -- CD247 GO:0050776 regulation of immune response TAS -- CD247 GO:0050852 T cell receptor signaling pathway TAS -- CLDN10 GO:0007155 cell adhesion TAS 10508613 CLDN10 GO:0016338 calcium-independent cell-cell adhesion ISS -- CRYBB3 GO:0007601 visual perception TAS 8999933 GEMIN2 GO:0000245 spliceosomal complex assembly IEA -- GEMIN2 GO:0000375 RNA splicing, via transesterification reactions TAS 9323129 GEMIN2 GO:0000387 spliceosomal snRNP assembly TAS -- GEMIN2 GO:0006397 mRNA processing TAS 9323130 GEMIN2 GO:0008380 RNA splicing TAS 9323129 GEMIN2 GO:0010467 gene expression TAS -- GEMIN2 GO:0016070 RNA metabolic process TAS -- GEMIN2 GO:0034660 ncRNA metabolic process TAS -- GRM1 GO:0000186 activation of MAPKK activity IEA -- GRM1 GO:0000187 activation of MAPK activity IEA -- GRM1 GO:0007186 G-protein coupled receptor signaling pathway TAS 7476890 GRM1 GO:0007268 synaptic transmission TAS 9076744 GRM1 GO:0007626 locomotory behavior IEA -- GRM1 GO:0008219 cell death IEA -- GRM1 GO:0019233 sensory perception of pain IEA -- GRM1 GO:0043408 regulation of MAPK cascade -- -- GRM1 GO:0071257 cellular response to electrical stimulus IEA -- Table S3 Cont. GRM1 GO:0051930 regulation of sensory perception of pain IEA -- NHLH2 GO:0006351 transcription, DNA-dependent IEA -- NHLH2 GO:0007417 central nervous system development TAS 1328219 NHLH2 GO:0030154 IEA -- NHLH2 GO:0045944 cell differentiation positive regulation of transcription from RNA polymerase II promoter TAS 16314316 NHLH2 GO:0007617 mating behavior IEA -- NHLH2 GO:0042698 ovulation cycle IEA -- NHLH2 GO:0051091 positive regulation of sequence-specific DNA binding transcription factor activity TAS 16314316 PALLD GO:0007010 cytoskeleton organization NAS 11598191 PCDHGC5 GO:0007156 homophilic cell adhesion IEA -- RASD1 GO:0007165 signal transduction TAS 10471929 RASD1 GO:0006913 nucleocytoplasmic transport -- -- RASD1 GO:0006184 GTP catabolic process -- -- RASD1 GO:0006886 intracellular protein transport -- -- RASD1 GO:0007186 TAS 10840027 RASD1 GO:0045892 G-protein coupled receptor signaling pathway negative regulation of transcription, DNAdependent IEA -- RASD1 GO:0007263 nitric oxide mediated signal transduction IEA -- RASD1 GO:0015031 protein transport -- -- RASD1 GO:0007264 small GTPase mediated signal transduction IEA -- RGL2 GO:0007264 small GTPase mediated signal transduction -- -- RGL2 GO:0007165 signal transduction -- -- RGL2 GO:0007265 Ras protein signal transduction NAS 8976381 RGL2 GO:0032318 NAS 8976381 RGL2 GO:0051056 regulation of Ras GTPase activity regulation of small GTPase mediated signal transduction -- -- RSAD2 GO:0001503 ossification IEA -- RSAD2 GO:0009615 IDA -- RSAD2 GO:0019048 response to virus modulation by virus of host morphology or physiology IEA -- RSAD2 GO:0030278 IEA -- RSAD2 GO:0034157 ISS -- RSAD2 GO:0034165 regulation of ossification positive regulation of toll-like receptor 7 signaling pathway positive regulation of toll-like receptor 9 signaling pathway ISS -- RSAD2 GO:0035710 CD4-positive, alpha-beta T cell activation ISS -- RSAD2 GO:0043367 CD4-positive, alpha-beta T cell differentiation ISS -- RSAD2 GO:0051607 defense response to virus IDA 16108059 RSAD2 GO:0045087 innate immune response IEA -- RSAD2 GO:0050709 negative regulation of protein secretion IDA 19074433 RSAD2 GO:0045071 IDA -- RSAD2 GO:2000553 negative regulation of viral genome replication positive regulation of T-helper 2 cell cytokine production ISS -- Table S3 Cont. SBK1 GO:0006468 protein phosphorylation -- -- SETD3 GO:0006351 transcription, DNA-dependent IEA -- SETD3 GO:0010452 histone H3-K36 methylation ISS -- SETD3 GO:0051568 histone H3-K4 methylation -- -- SETD3 GO:0018027 peptidyl-lysine dimethylation ISS -- SETD3 GO:0018026 peptidyl-lysine monomethylation ISS -- SETD3 GO:0018023 peptidyl-lysine trimethylation ISS -- SETD3 GO:0051149 IEA -- SETD3 GO:0045944 IEA -- SETD3 GO:0045893 positive regulation of muscle cell differentiation positive regulation of transcription from RNA polymerase II promoter positive regulation of transcription, DNAdependent ISS -- STYK1 GO:0006468 protein phosphorylation -- -- TMEM150A GO:0009056 catabolic process IEA -- TUBA1A GO:0051084 'de novo' posttranslational protein folding TAS -- TUBA1A GO:0000086 G2/M transition of mitotic cell cycle TAS -- TUBA1A GO:0000226 microtubule cytoskeleton organization -- -- TUBA1A GO:0000278 mitotic cell cycle TAS -- TUBA1A GO:0006184 GTP catabolic process -- -- TUBA1A GO:0006457 protein folding TAS -- TUBA1A GO:0051301 cell division TAS 12090300 TUBA1A GO:0044267 cellular protein metabolic process TAS -- TUBA1A GO:0030705 cytoskeleton-dependent intracellular transport TAS 12090300 TUBA1A GO:0007017 microtubule-based process TAS 12090300 TUBA1A GO:0051258 protein polymerization IEA -- Evidence1 PubMed Reference Molecular Function Gene Symbol ID GO term AATK GO:0004674 protein serine/threonine kinase activity IEA -- AATK GO:0005524 ATP binding IEA -- AATK GO:0004672 protein kinase activity -- -- AATK GO:0004713 protein tyrosine kinase activity AATK GO:0005515 AATK GO:0016772 protein binding transferase activity, transferring phosphoruscontaining groups ADHFE1 GO:0016491 oxidoreductase activity ADHFE1 GO:0046872 metal ion binding IEA -- ADHFE1 GO:0047988 hydroxyacid-oxoacid transhydrogenase activity IDA 16435184 ADORA3 GO:0001609 G-protein coupled adenosine receptor activity IEA -- ADORA3 GO:0004930 -- -- AHR GO:0003700 G-protein coupled receptor activity sequence-specific DNA binding transcription factor activity IDA 11782478 IEA -- IPI 14521924 -- -- -- -- Table S3 Cont. AHR GO:0003677 DNA binding RNA polymerase II distal enhancer sequencespecific DNA binding transcription factor activity TAS 19538249 AHR GO:0003705 IEA -- AHR GO:0004871 AHR GO:0004879 signal transducer activity ligand-activated sequence-specific DNA binding RNA polymerase II transcription factor activity -- -- AHR GO:0008134 transcription factor binding IDA 10395741 IPI 9079689 AHR GO:0044212 transcription regulatory region DNA binding IDA 15681594 AHR GO:0051879 Hsp90 protein binding IDA 9079689 AHR GO:0043565 sequence-specific DNA binding IEA -- AHR GO:0046983 protein dimerization activity TAS 19538249 AHR GO:0046982 protein heterodimerization activity TAS 19538249 AHR GO:0005515 protein binding IPI 10395741 CD247 GO:0004888 transmembrane signaling receptor activity IEA -- CD247 GO:0005515 protein binding IPI 10704231 CD247 GO:0042802 identical protein binding IPI 17055436 CD247 GO:0042803 protein homodimerization activity NAS 12110186 CLDN10 GO:0005198 structural molecule activity IEA -- CLDN10 GO:0042802 identical protein binding ISS -- CRYBB3 GO:0005212 structural constituent of eye lens IEA -- DDX10 GO:0005524 ATP binding IEA -- DDX10 GO:0008026 ATP-dependent helicase activity IEA -- DDX10 GO:0004386 helicase activity -- -- DDX10 GO:0003676 nucleic acid binding -- -- DDX10 GO:0003723 RNA binding IEA -- DDX10 GO:0003724 RNA helicase activity TAS 8660968 GEMIN2 GO:0005515 protein binding IPI 11714716 GRM1 GO:0004930 G-protein coupled receptor activity TAS 7476890 GRM1 GO:0005515 protein binding IPI 19084525 GRM1 GO:0008066 glutamate receptor activity TAS 7476890 HES6 GO:0003677 IEA -- HES6 GO:0003700 DNA binding sequence-specific DNA binding transcription factor activity IEA -- HES6 GO:0046983 protein dimerization activity IEA -- HES6 GO:0003712 transcription cofactor activity ISS 10851137 HES6 GO:0008134 transcription factor binding NAS 17611704 MYOM3 GO:0005515 protein binding -- -- MYOM3 GO:0042803 protein homodimerization activity ISS -- NEUROD4 GO:0003677 DNA binding IEA -- NEUROD4 GO:0046983 protein dimerization activity IEA -- Table S3 Cont. NHLH2 GO:0001102 RNA polymerase II activating transcription factor binding NHLH2 GO:0003677 NHLH2 IPI 16314316 DNA binding IEA -- GO:0046983 protein dimerization activity IEA -- NHLH2 GO:0005515 protein binding IPI -- PALLD GO:0003779 actin binding IEA -- PALLD GO:0005515 protein binding IPI 11598191 PALLD GO:0051371 muscle alpha-actinin binding TAS 16125169 PCDHGC5 GO:0005509 calcium ion binding IEA -- RASD1 GO:0005525 GTP binding IEA -- RASD1 GO:0003924 GTPase activity IEA -- RASD1 GO:0005515 protein binding -- -- RGL2 GO:0005088 Ras guanyl-nucleotide exchange factor activity NAS 8976381 RGL2 GO:0005085 guanyl-nucleotide exchange factor activity -- -- RSAD2 GO:0003674 molecular_function ND -- RSAD2 GO:0003824 catalytic activity IEA -- RSAD2 GO:0005515 protein binding IPI -- RSAD2 GO:0043621 protein self-association IDA 19074433 RSAD2 GO:0046872 metal ion binding IEA -- RSAD2 GO:0051536 iron-sulfur cluster binding -- -- RSAD2 GO:0051539 4 iron, 4 sulfur cluster binding IDA -- SBK1 GO:0004672 protein kinase activity -- -- SBK1 GO:0005524 ATP binding IEA -- SBK1 GO:0004674 protein serine/threonine kinase activity IEA -- SBK1 GO:0004713 -- -- SBK1 GO:0016772 -- -- SETD3 GO:0001102 protein tyrosine kinase activity transferase activity, transferring phosphoruscontaining groups RNA polymerase II activating transcription factor binding IEA -- SETD3 GO:0003713 ISS -- SETD3 GO:0046975 ISS -- SETD3 GO:0042800 transcription coactivator activity histone methyltransferase activity (H3-K36 specific) histone methyltransferase activity (H3-K4 specific) IEA -- SETD3 GO:0005515 protein binding -- -- STYK1 GO:0004672 protein kinase activity -- -- STYK1 GO:0004713 protein tyrosine kinase activity -- -- STYK1 GO:0005524 IEA -- STYK1 GO:0004715 ATP binding non-membrane spanning protein tyrosine kinase activity IEA -- STYK1 GO:0005515 IPI -- STYK1 GO:0016772 -- -- protein binding transferase activity, transferring phosphoruscontaining groups Table S3 Cont. TUBA1A GO:0003924 GTPase activity IEA -- TUBA1A GO:0005198 structural molecule activity TAS 12090300 TUBA1A GO:0005200 structural constituent of cytoskeleton IEA -- TUBA1A GO:0005515 protein binding IPI 19893491 TUBA1A GO:0005525 GTP binding IEA -- TUBA1A GO:0019904 protein domain specific binding IEA -- Evidence1 PubMed Reference Cellular Component Gene Symbol ID GO term AATK GO:0005783 endoplasmic reticulum IEA -- AATK GO:0048471 perinuclear region of cytoplasm IEA -- AATK GO:0005739 mitochondrion -- -- AATK GO:0016021 integral component of membrane IEA -- AATK GO:0044295 axonal growth cone IEA -- AATK GO:0055037 recycling endosome IEA -- ADHFE1 GO:0005739 mitochondrion ISS -- ADHFE1 GO:0005759 mitochondrial matrix TAS -- ADORA3 GO:0005886 plasma membrane TAS -- ADORA3 GO:0005887 integral component of plasma membrane IEA -- ADORA3 GO:0016021 integral component of membrane IEA -- ADORA3 GO:0042629 mast cell granule IEA -- AHR GO:0005667 transcription factor complex TAS 19538249 AHR GO:0005634 nucleus IDA -- AHR GO:0005730 NOT nucleolus IDA -- AHR GO:0005737 cytoplasm IDA -- AHR GO:0034752 cytosolic aryl hydrocarbon receptor complex TAS 19538249 CD247 GO:0005737 cytoplasm IDA 11390434 CD247 GO:0005886 plasma membrane TAS -- CD247 GO:0016020 membrane -- -- CD247 GO:0016021 integral to membrane IEA -- CD247 GO:0042101 T cell receptor complex IDA 8176201 CD247 GO:0042105 alpha-beta T cell receptor complex IEA -- CLDN10 GO:0005886 plasma membrane IEA -- CLDN10 GO:0005923 tight junction ISS -- CLDN10 GO:0016020 membrane -- -- CLDN10 GO:0016021 integral to membrane IEA -- GEMIN2 GO:0005622 intracellular -- -- GEMIN2 GO:0005634 nucleus TAS 9323130 Table S3 Cont. GEMIN2 GO:0005654 nucleoplasm TAS -- GEMIN2 GO:0005681 spliceosomal complex IEA -- GEMIN2 GO:0005737 cytoplasm TAS 9323130 GEMIN2 GO:0005829 cytosol TAS -- GEMIN2 GO:0015030 Cajal body -- -- GEMIN2 GO:0097504 Gemini of coiled bodies IEA -- GRM1 GO:0005634 nucleus IEA -- GRM1 GO:0005886 plasma membrane TAS -- GRM1 GO:0005887 integral to plasma membrane TAS 7476890 GRM1 GO:0014069 postsynaptic density IEA -- GRM1 GO:0016021 integral to membrane -- -- GRM1 GO:0030425 IEA -- GRM1 GO:0038038 dendrite G-protein coupled receptor homodimeric complex IPI 10945991 GRM1 GO:0043005 neuron projection -- -- HES6 GO:0005634 nucleus IEA -- HES6 GO:0005667 transcription factor complex ISS 10851137 MYOM3 GO:0031430 M band ISS -- NEUROD4 GO:0005634 nucleus IEA -- NHLH2 GO:0005634 nucleus IEA -- NHLH2 GO:0005667 transcription factor complex TAS 16314316 PALLD GO:0001726 ruffle IEA -- PALLD GO:0005634 nucleus IDA 11598191 PALLD GO:0005884 actin filament IDA 11598191 PALLD GO:0005925 focal adhesion IEA -- PALLD GO:0030018 Z disc IEA -- PCDHGC5 GO:0005886 plasma membrane IEA -- PCDHGC5 GO:0016020 membrane -- -- PCDHGC5 GO:0016021 integral to membrane IEA -- RASD1 GO:0016020 membrane -- -- RASD1 GO:0005886 plasma membrane IEA -- RASD1 GO:0005622 intracellular -- -- RASD1 GO:0005634 nucleus IEA -- RASD1 GO:0048471 perinuclear region of cytoplasm IEA -- RGL2 GO:0005622 intracellular IEA -- RGL2 GO:0005575 cellular_component ND -- RSAD2 GO:0005739 mitochondrion IDA -- RSAD2 GO:0005741 mitochondrial outer membrane IDA -- RSAD2 GO:0005743 mitochondrial inner membrane IDA -- Table S3 Cont. RSAD2 GO:0005783 endoplasmic reticulum IDA 11752458 RSAD2 GO:0005789 endoplasmic reticulum membrane IDA 19074433 RSAD2 GO:0005794 Golgi apparatus IEA -- RSAD2 GO:0005811 lipid particle ISS -- SBK1 GO:0005737 cytoplasm IEA -- SETD3 GO:0000790 nuclear chromatin IEA -- STYK1 GO:0005886 plasma membrane IDA -- STYK1 GO:0016021 integral to membrane IEA -- TMEM150A GO:0005886 plasma membrane IEA -- TMEM150A GO:0016021 integral component of membrane IEA -- TUBA1A GO:0005829 cytosol TAS -- TUBA1A GO:0005874 microtubule TAS 12090300 TUBA1A GO:0005881 cytoplasmic microtubule IEA -- TUBA1A GO:0015630 microtubule cytoskeleton IDA -- TUBA1A GO:0043234 protein complex -- -- 1 Inference Codes: IDA = Direct Assay; IEA = Electronic Annotation; IPI = Physical Interaction; ISS = Sequence or structural Similarity; NAS = Non-traceable Author Statement; ND = No biological Data available; TAS = Traceable Author Statement Table S4. Significant canonical pathways, functions, and top networks for the 26 annotated DMRs identified using Ingenuity Pathway Analysis. Canonical Pathways Name Sertoli Cell-Sertoli Cell Junction Signaling Role of Lipids/Lipid Rafts in the Pathogenesis of Influenza Cytotoxic T Lymphocyte-mediated Apoptosis of Target Cells Ethanol Degradation II G-Protein Coupled Receptor Signaling p-value 2.1E-02 3.1E-02 3.9E-02 4.1E-02 4.1E-02 Molecular and Cellular Functions Name Cellular Development p-value 2.5E-04 - 4.4E-02 Cell Morphology 4.1E-04 - 4.1E-02 Cell-To-Cell Signaling and Interaction Amino Acid Metabolism Cell Cycle 9.3E-04 - 3.9E-02 1.3E-03 - 4.5E-02 1.3E-03 - 3.2E-02 # of Genes Included DMR Genes 13 AATK, ADORA3, AHR, CD247, GEMIN2, HES6, NEUROD4, PALLD, RASD1, RGL2, RSAD2, SETD3, TUBA1A 11 AATK, ADORA3, AHR, CD247, GRM1, KIAA1467, NEUROD4, NHLH2, PALLD, PCDHGC5, TUBA1A 6 ADORA3, AHR, CD247, GRM1, PCDHGC5, PALLD 2 GRM1, SBK1 4 AHR, CD247, RGL2, TUBA1A Physiological System Development and Function Name Organ Morphology Organismal Development Reproductive System Development and Function Nervous System Development and Function Auditory and Vestibular System p-value 1.9E-04 - 4.3E-02 1.9E-04 - 4.7E-02 1.9E-04 - 4.2E-02 2.0E-04 - 4.5E-02 1.30E-03 # of Genes Included DMR Genes 3 AHR, CD247, NHLH2, 2 ADORA3, NHLH2 4 AHR, CD247, GRM1, NHLH2 7 AATK, CD247, GRM1, NEUROD4, NHLH2, PALLD, TUBA1A 1 GRM1 Development and Function Top Networks Associated Network Functions Cellular Assembly and Organization, Cellular Compromise, Nervous System Development and Function Nervous System Development and Function, Organ Morphology, Organismal Development ID_Score 1_38 2_26 # of Genes Included DMR Genes 15 AATK, ADHFE1, CCDC93, CLDN10, CRYBB3, DDX10, GEMIN2, HES6, KIAA1467, MYOM3, PALLD, PCDHGC5, RGL2, SETD3, TMEM150A 11 ADORA3, AHR, CD247, GRM1, NEUROD4, NHLH2, RASD1, RSAD2, SBK1, STYK1, TUBA1A