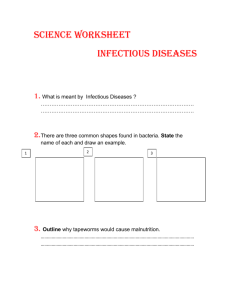
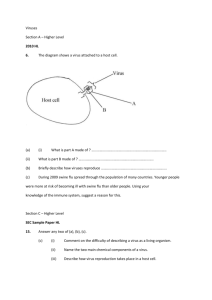
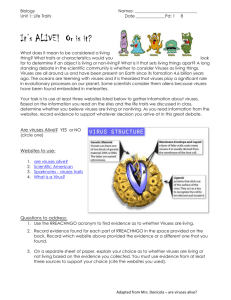
Project Title: The Evolution of Severity in Viruses
advertisement

Masters Student Bursary Available (July 2013) Project Title: The Evolution of Severity in Viruses Hosts/Supervisors: This is a joint collaboration between the South African National Bioinformatics Institute (SANBI) at the University of the Western Cape, the Department of Molecular and Cell Biology (MCB) at the University of Cape Town (UCT), and the Computational Biology Group (CBIO) at the Institute of Infectious Disease and Molecular Medicine (IIDMM) at UCT. Supervisors are Dr Gordon Harkins (SANBI), Dr Dionne Shepherd (MCB) and Dr Darren Martin (CBIO). Wet bench work will be done at MCB, with computational/bioinformatics support given by SANBI and CBIO. Requirements: BSc (Hons) in any subject that has provided exposure to molecular biology techniques. Start Date: July 2013. Project Summary: We are hoping to recruit an enthusiastic, intelligent and hard-working masters student and are offering a two year scholarship (R55 000 per year) to work on a cutting edge computational and wet-laboratory based project dealing with the evolution of virus severity. Almost all virus diseases we know of originated when a virus from one species began infecting the members of a different species. Well known examples of this include swine flu from a few years ago or HIV (which began infecting humans from chimpanzees about 120 years ago). It is widely believed that when a virus begins infecting a new host species, the virus initially will have a sub-optimal degree of severity in its new host. What this means is that it will either be so severe that it kills the host before the host is able to efficiently transmit it to different members of its species, or that it will be so mild that not enough viruses are transmitted to initiate new infections in new individuals. Over time such viruses are expected to evolve to maximise their chances of being transmitted by becoming either more or less severe. Knowing where any particular group of viruses sits on this journey between lower and higher severity (and which of these is their destination) is extremely interesting to scientists that are trying to forecast future trends in virus evolution and epidemiology. Unfortunately it is not possible to physically go back in time and measure the severities of ancient viruses. Luckily, however, modern computational and laboratory tools have been developed that now make it possible for us to estimate how severe long extinct viruses were. Specifically, based on both the evolutionary relatedness and measurable severities of viruses that are currently in existence, newly developed computational tools can approximate the severities of their ancient ancestors. It is, however, not presently known how accurate these new methods are. Based on the full genome nucleotide sequences of contemporary viruses it is possible to infer the genomic nucleotide sequences of these same ancestral viruses. This is really important because modern advances in DNA synthesis and the artificial reconstruction of “living” viruses means that it is now possible to actually recreate these ancestral viruses and directly test how severe they actually were. Although carrying out this work on human or domesticated animal viruses would be unethical and extremely dangerous, there are no such issues with plant viruses. The project will therefore focus on a model plant-infecting virus that causes maize streak disease. Besides having an extremely small DNA genome and easily measurable severity-related biological characteristics (i.e. it is very easy to work with both computationally and in the lab), maize streak virus (the virus species that causes maize streak disease) evolves as rapidly as most human and animal viruses and is therefore an ideal subject for this project. The student will work with a research group that includes computational and wet-lab scientists working together at the cutting-edge of an exciting new research area. They will initially be involved in inferring and synthesising full genome sequences of long extinct viruses and will then determine how closely the computationally inferred biological characteristics of these viruses match their actually measured characteristics. Besides addressing an important unanswered question in modern evolutionary biology, the student will be exposed to a wide array of modern computational and laboratory-based approaches that will empower them to tackle a wide range of molecular-evolution and epidemiology based projects. Contact numbers for further inquiries: Dr. Harkins: 021 959 2096; 082 596 1794 Dr. Shepherd: 021 650 5232; 084 200 1886 Applications, including CVs and competencies, should be sent to gordon@sanbi.ac.za or dionne.shepherd@gmail.com