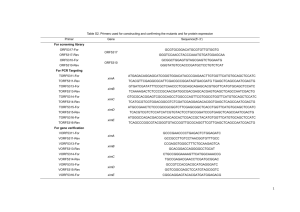
Table S1. Catabolic and transport genes that are up
advertisement

Table S1. Catabolic and transport genes that are up-regulated in the absence of Hfq ORF or operona PA0119 Gene PA0129 Functiona Predicted Hfq binding motifsc putative dicarboxylate transporter Fold change hfqvs wtb 10 bauD amino acid permease 4.5 PA0153PA0154 PA0162 pcaHG protocatechuate degradation ATAACGCGAATAACAGGAGCCCGACatg (pcaH) opdC histidine porin OpdC 9.7 (pcaH), 4.0 (pcaG) 3.6 PA0226PA0228 PA0297PA0299 PA0300 pcaF protocatechuate degradation 11 spuABC glutamine biosynthesis spuD polyamine transport protein 7.6 (spuA), 5.6 (spuB), 6.4 (spuC) 4.5 ACAACAAGACGAGACCCGACCGatg (PA0226) CAGAAGGGGAGCGACGAatg (pcaF) ATAACATCTACAAACAACGGGTGTCTCatg (spuA); AATAGAAGAGGTGTGACatg (spuC) CAACAAAAATGGAGCTACCCCGCatg PA0301PA0304 spuEFGH polyamine transport proteins AAAAGGCCCGGAACAGGAGTCGGACatg (spuE) AACAGCCTGAAAAAGCGCatg (spuG) AAAGAACTGGAGGGCAAGGTatg (spuH) AAGAAACCAGAAGAGAGGCTACAACCgtg (PA0440) dht putative oxidoreductases for glutamate biosynthesis dihydropyrimidinase 4.8 (spuE), 7.8 (spuF), 7.3 (spuG), 5.7 (spuH) 129 (-39), 99 (-40) 45 PA0443PA0444 PA0783 95 (-43), 11 (-44) putP putative transporter and N-carbamoyl-betaalanine amidohydrolase sodium/proline symporter AAAAAACAATTTCAAGAACCGGCAACGACCGGTCAGCCTGCGAGGAAA GACGGCatg AATAACTAAAAGAATTCTGGAGAGGCCCatgCAACAGAGCAGA (PA0443) 3.6 ATAAAAACAAGCAATAGGGGAGTCCCTatg PA0866 aroP2 aromatic amino acid transport protein 84 ACAACAATAAATTTCCACTTGCGTGAGATAATTCAatg PA0870PA0872 PA1019 phhABC phenylalanine metabolism mucK cis.cis-muconate transporter 2.5 (phhB), 2.3 (phhC) 3.2 AACAACAAAGACCCCGCGGCCACAAGCCGCTCATGGAGTCCGTatg (phhA) AGAACAAGAATAatg PA1071PA1073 braDEFG branched-chain amino acid transport proteins CCGGAGGTGGAAAAAGTatg (braE) PA1074 braC branched-chain amino acid transport protein 9.9 (braD), 3.2 (braF), 8.0 (braE), 4.3 (braG) 3.2 PA1338 ggt gamma-glutamyltranspeptidase precursor 11 GATAACAATAAATAGGGAAAGCCCatg PA1763 hypothetical protein 6.6 AAAAACAAGAACAATCTCTGGGGATAGCGCCatg PA1891PA1897 hypothetical proteins TAAGGAAGAGGAGATAAATatg (PA1897) PA0439PA0440 PA0441 PA1978 erbR glycerol metabolism activator 13 (-91), 9.6 (-92), 18 (-93), 19 (-94), 19 (-95), 31 (-96), 36 (-97) 22 PA1979PA1980 eraSR two component regulator for quinoprotein ethanol dehydrogenase 5.7 (eraS), 7.2 (eraR) AATAAAAATGCGTTCATCGCCCGCCTCCGCGAGGCCCGATGGAAGCGC CGAatg AAGAACAATTAAAGGGTCTCAACGatg AAGAAGGAGCAACCGCAatgAGGAAT ACAACAATGACAACACACAAGAAGAGTGGAGCACTatgAAGAAG CATAACGACAATAACGAGGTGCAGGCCatg PA1981PA1982 PA1983PA1984 PA1991PA1992 PA2003 exaA quinoprotein ethanol dehydrogenase 29 (-81), 58 (exaA) atgACAACAAGAAC (exaA) exaBC ethanol utilization AGGAATGAAGTGatgAACAAGAACAAC (exaB) ercS ethanol utilization 12 (exaB), 12 (exaC) 5.6 (-91), 5.1 (ercS) bdhA 3-hydroxybutyrate dehydrogenase 6.1 AGAAACAGCatg 13 AATAAGAACCTCAGGTACCTTGCGatg PA2079 probable major facilitator superfamily (MFS) transporter probable amino acid permease 6.0 AAGAACCCCAATAAGATTCCAGAGGACATCCAGatgAAC PA2210 probable MFS transporter 21 AAGAACAACAATAAAAGAGGTACGACCCatg branched chain aminoacid assimilation 3.2 (bkdA1), 6.2 (bkdA2), 6.0 (bkdB), 4.1 (lpdV) 35 CAACGCCAGGAGCTCGGGGTatg (bkdA2) 4.1 (-64), 3.8 (-65), 5.5 (-66) 197 (catB), 87 (catC), 98 (catA) 11 AGAAAGACGAAACCGTGATGATCCAGAGAGGACCCCACCatg (PA2264) PA2006 PA2247PA2250 bkdA1A2B lpdV PA2262 putative 2-ketogluconate transporter PA2264PA2266 PA2507PA2509 PA2511 gluconate dehydrogenase (PA2265) AACAATAAGAATGGTCTGTCAGatg (PA1991) ACAAGAACAAGACAAGCTCAGGAGATACCCGACCatg catBCA catechol degradation antR transcriptional regulator PA2512PA2514 PA2515PA2518 antABC anthranilate degradation xylXYZL toluate degradation PA2519 xylS transcriptional regulator 80 (antA), 67 (antB), 152 (antC) 16 (xylX), 6.6 (xylY), 3.9 (xylZ), 7.2 (xylL) 15 PA2760 oprQ outer membrane protein Q 5.0 AACAACCAGGAACAATAAGatgTTGAAGAAAAGG PA3186 oprB glucose/carbohydrate outer membrane porin 5.7 AAatgTACAAGAACAAGAAAACCAGA PA3366 amiEBCRS aliphatic amidase PA3569PA3570 PA3709 mmsAB branched chain aminoacid assimilation 7.2 (amiE), 12 (amiB), 4.7 (amiC), 4.7 (amiR), 2.1 (amiS) 5.0 (mmsA), 7.3 (mmsB) 30 AAAAAATAACAACAAGAGGTGATATCCatg (amiE) CAGAAGGAGTTTCATCCatg (amiB) AGCGACAGTCACAGGAGAGGAAACGGatg (amiC) AACAATAACAAGAGGGGTATCGTCatg (amiS) AACAATAAAAGCCGGCCTGGCCGGTGCAGGAGGGTTCatg (mmsA) PA4023PA4025 PA4091PA4092 PA4121PA4123 PA4124PA4125 eutPBC ethanolamine utilization GATAACAACAAGATTGAGGTGAAAAACCCatg (eutP); hpaAC hydroxyphenylacetate degradation hpcC hydroxyphenylacetate degradation hpcBD hydroxyphenylacetate degradation 57 (eutP), 4.5 (eutB), 2.1 (eutC) 16 (hpaA), 9.8 (hpaC) 29 (-21), 20 (-22), 90 (-23) 12 (hpcB), 17 (hpcD) probable MFS transporter GACAACAACAAGCAGAGGCAAACGCCatg (catA) CAGACAGAGGAGAAACGACCGatg (catC) AAAACAGAGCGCCAGCAGAGCGCACCCCCAGCCTTGCCGTGTCGAGTG CCGAACCatgATGAGG CATAACGACAACGCACAAGGGTGAGAACGCatg (antA) GAACAATAACAACGGAGGCCCCGGCCatg (xylX) ATAACAACAACGATAGCAGCCTGAGTGTCCTGCCCatg (xylS) AGACAACAATAAGAGCCCCCACAAGGGGCGGGAGAGCCGCAatg AGATAACAACAATGACTGGAGACGATGatg (hpaA) CCAGGAGACACCACCACAatg (PA4121) AATGAGGAACCGACCatg (PA4122) PA4126PA4128 PA4137 hpcG PA4209 phzM CACAAAAAGTACAAGATCatg (PA4126) putative porin 8.5 (-26), 11 (hpcC), 11 (-28) 989 phenanzine specific methyltransferase 37 AACGAGAGAGAATAAAAGatgAATAATTCGAAT 4.7 AGACAATCAAAATAACCAGAGGTCACTCCatg PA4498 putative binding protein component of ABC transporter putative metallopeptidase 3.8 AGACGAGGAGAACAACAAatg PA4500 putative ABC transporter protein 19 TAGAAAAAGAAAAAATGAGGTTTGCatg 77 (opdD), 28 (02), 15 (-03), 20 (04), 11 (-05), 11 (06) 6.5 AACGACAACAATAAAGGGAGCAATCAGgtg (opdD) 8.0 (-08), 16 (-09), 24 (-10), 20 (-11), 53 (-12) 26 GAAGAGACTGAAAAatg (PA4912) AAGAGGAGCTGTCatg (PA4909) PA4496 hydroxyphenylacetate degradation PA4501PA4506 opdD glycine-glutamate dipeptide uptake PA4588 gdhA glutamate dehydrogenase PA4908PA4912 putative ABC transporter proteins PA4913 probable binding protein component of ABC transporter probable oxidoreductase PA4986 PA5112 estA CAAAAAATTGTTCACAAAACTAGAAACCGGTGAGAAAAatg 32 AAACACAACAACAACATCGTAGTTACGAGCGCCAACATCATCCTCAGG AGCAACACCCCatg AACAACAAGAAGGACGCCAACCgtg esterase 2.8 GACAATAAAAACAAATCATGGAGTAAGAGAatg putative permease of ABC transporter 2.9 (-52), 5.0 (-53), 4.9 (-54), 3.2 (-55) 3.5 AATAACAAGAGGAAAATCCatg (PA5152 first gene in operon) 7.4 AACAATCATAAAAAAAGCCTCGTGAGAGGCCTGCTTGCTCAGCGTGAG GAGATACACCCatg atgGCCCGCGAGCAGCAAC (PA5523) PA5152PA5155 PA5309 pauB4 FAD-dependent oxidoreductase PA5380 gbdR PA5522PA5523 PA5542 pauA6 Transcriptional regulator for glycine betaine catabolism glutamylpolyamine synthetase PA5543PA5545 AAGAACAACAACCAAGGGAAGAATCGatg hypothetical proteins hypothetical proteins 3.2 (-22), 4.3 (pauA6) 7.6 12 (-45), 30 (-44), 8.3 (-43) AATAATCAGGATTTCACACatg CATAAGGAAAACGTCCatg GACAACAACGATAAGAACAGGAGACTTCCCatg (PA5545) a Gene numbers and functions are taken from the Pseudomonas genome database [61] b data derived from Sonnleitner et al [31] c A-rich sequences are highlighted in bold; start codons are in lower case letters.