DNA methylation patterns in ANCA Associated Vasculitis
advertisement
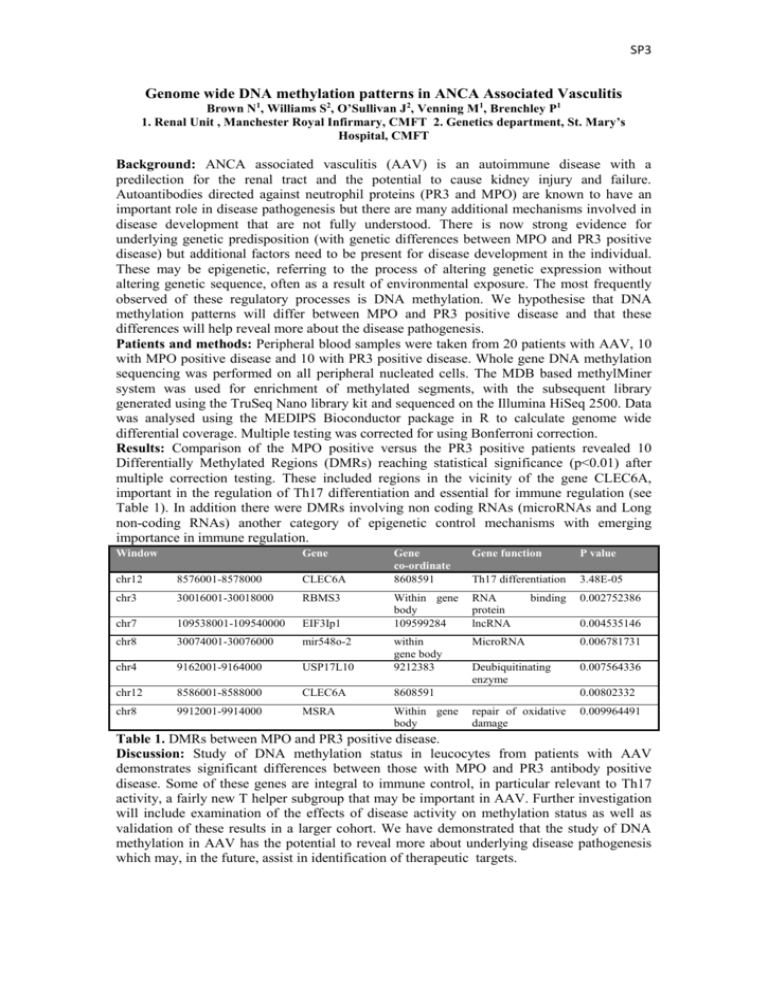
SP3 Genome wide DNA methylation patterns in ANCA Associated Vasculitis Brown N1, Williams S2, O’Sullivan J2, Venning M1, Brenchley P1 1. Renal Unit , Manchester Royal Infirmary, CMFT 2. Genetics department, St. Mary’s Hospital, CMFT Background: ANCA associated vasculitis (AAV) is an autoimmune disease with a predilection for the renal tract and the potential to cause kidney injury and failure. Autoantibodies directed against neutrophil proteins (PR3 and MPO) are known to have an important role in disease pathogenesis but there are many additional mechanisms involved in disease development that are not fully understood. There is now strong evidence for underlying genetic predisposition (with genetic differences between MPO and PR3 positive disease) but additional factors need to be present for disease development in the individual. These may be epigenetic, referring to the process of altering genetic expression without altering genetic sequence, often as a result of environmental exposure. The most frequently observed of these regulatory processes is DNA methylation. We hypothesise that DNA methylation patterns will differ between MPO and PR3 positive disease and that these differences will help reveal more about the disease pathogenesis. Patients and methods: Peripheral blood samples were taken from 20 patients with AAV, 10 with MPO positive disease and 10 with PR3 positive disease. Whole gene DNA methylation sequencing was performed on all peripheral nucleated cells. The MDB based methylMiner system was used for enrichment of methylated segments, with the subsequent library generated using the TruSeq Nano library kit and sequenced on the Illumina HiSeq 2500. Data was analysed using the MEDIPS Bioconductor package in R to calculate genome wide differential coverage. Multiple testing was corrected for using Bonferroni correction. Results: Comparison of the MPO positive versus the PR3 positive patients revealed 10 Differentially Methylated Regions (DMRs) reaching statistical significance (p<0.01) after multiple correction testing. These included regions in the vicinity of the gene CLEC6A, important in the regulation of Th17 differentiation and essential for immune regulation (see Table 1). In addition there were DMRs involving non coding RNAs (microRNAs and Long non-coding RNAs) another category of epigenetic control mechanisms with emerging importance in immune regulation. Window Gene Gene co-ordinate 8608591 Gene function P value Th17 differentiation 3.48E-05 Within gene body 109599284 RNA protein lncRNA 0.002752386 MicroRNA 0.006781731 Deubiquitinating enzyme 0.007564336 chr12 8576001-8578000 CLEC6A chr3 30016001-30018000 RBMS3 chr7 109538001-109540000 EIF3Ip1 chr8 30074001-30076000 mir548o-2 chr4 9162001-9164000 USP17L10 within gene body 9212383 chr12 8586001-8588000 CLEC6A 8608591 chr8 9912001-9914000 MSRA Within gene body binding 0.004535146 0.00802332 repair of oxidative damage 0.009964491 Table 1. DMRs between MPO and PR3 positive disease. Discussion: Study of DNA methylation status in leucocytes from patients with AAV demonstrates significant differences between those with MPO and PR3 antibody positive disease. Some of these genes are integral to immune control, in particular relevant to Th17 activity, a fairly new T helper subgroup that may be important in AAV. Further investigation will include examination of the effects of disease activity on methylation status as well as validation of these results in a larger cohort. We have demonstrated that the study of DNA methylation in AAV has the potential to reveal more about underlying disease pathogenesis which may, in the future, assist in identification of therapeutic targets.