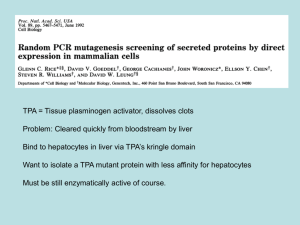
tpj12844-sup-0002-Legends
advertisement

Supporting Information Legends Figure S1: Protein blot analysis of the efficiency of TaV 2A processing in Chlamydomonas cells. Proteins from cells transformed with mCherry Bler-TaV 2AmCherry-CIA5 Ep gene construct detected on a protein blot using polyclonal antibodies raised against a synthetic CIA5 epitope (CIA5 Ep). Lane1) A zeocin resistant (Bler) culture not expressing mCherry. Lanes 2 and 3) Zeocin resistant cultures expressing and processing Bler-TaV 2A-mCherry-CIA5 Ep. Expected sizes of indicated bands are as follows: CIA5 = ~100 kDa, full length unprocessed Bler- TaV 2A-mCherry-CIA5 Ep = 48.6 kDa, mCherry-CIA5 Ep = 30.4 kDa. Figure S2: FKB12 complementation assay. A) Zeocin resistant colonies of originally rapamycin-resistant Chlamydomonas fkb12 mutants transformed with plasmid pMA4 containing a nonfunctional FMDV 2A coding sequence separating a zeocin resistance gene and a FKB12 gene (left column) or a separate set of colonies generated from cells transformed with plasmid pLZ1 containing a functional FMDV 2A coding sequence separating a zeocin resistance gene and a FKB12 gene (right column). Colonies were randomly picked to TAP liquid medium in microtiter plates, incubated overnight, and replica plated onto TAP plates (top row) and TAP plates containing rapamycin (bottom row). Successful complementation of fkb12 mutants with a functional FKB12 gene results in loss of rapamycin resistance. B) A second fkb12 mutant containing a 21 bp deletion and a 4 bp insertion in the FKB12 gene (bottom DNA sequence) was transformed with pLZ1 and a set of the resultant zeocin resistant colonies of the originally rapamycin-resistant Chlamydomonas mutants were randomly picked to TAP liquid medium in microtiter plates, incubated overnight, and replica plated onto TAP plates (left) and TAP plates containing rapamycin (right). WT, wild type. Figure S3: Enlargement of portions of Figure 6A to better visualize mCherry-tagged Rubisco in the Chloroplast of cells transformed with plasmid QN4. A) Portions of original image of cells maintained in high CO2 conditions (lower left panel) as contrasted to the exclusive localization of mCherry-tagged Rubisco in the pyrenoid of the same cells maintained in low CO2 concentrations (upper left panel). B) Red color enhancement with Photoshop of images shown in A to more easily visualize the exclusive localization of mCherry-tagged Rubisco fluorescence to the pyrenoid (upper left panel) and the patterns of mCherry-Rubisco redistribution to the chloroplast stroma following shift of cells from low CO 2 to high CO2 conditions (lower left panel). Figure S4: Evaluation of various selectable markers for compatibility with the extended FMDV 2A peptide. A) Constructs for testing effects of opposite orientations of a gene of interest (GOI) and selectable markers in 2A-containing dicistronic genes. Gluc, Gaussia Luciferase; 2A, extended FMDV 2A peptide; PsaD Pro, promoter from Chlamydomonas PsaD gene. B) Performance of each construct in regard to effect on transformation rates, maintenance or loss of linkage between the GOI (Gluc) and the selectable marker gene, and the approximate relative luminescence units (RLUs) measured in cultures of at least 10 randomly chosen transformants. Linkage was defined as detection of Gluc signal in at least 70% of transformants tested. Bler (Zeocinr); Aph8 (Paromomycinr); Aph7 (Hygromycinr); AadA (Spectinomycinr); Arg7 (argininosuccinate lyase; complementation of arginine auxotrophy in arg7 mutants); RbcS2 (Rubisco small subunit 2, complementation of photoautotrophy in rbcs null mutants); pSP124, standard laboratory construct containing a Bler gene driven by the RbcS2 gene promoter (used as a control).