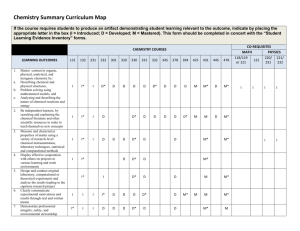
The Semantification of Chemistry
Author(s):
Lezan Hawizy
Executive Summary
Chemistry is a central science and the data produced as a consequence is immense. However, much of
this data is which makes data integration difficult. In this article, we demonstrate how chemical data
can be retrieved from reports, scientific theses and papers or patents and discuss how these sources
can be processed using natural language processing techniques and named-entity recognisers to
produce chemical data and knowledge expressed in RDF.
1. Introduction
Chemistry is at the heart of several high-value industries, such as the pharmaceutical and biomedical
industries, materials manufacturing and design etc. Future progress in science will crucially depend on
the ability to mash up chemical data with data derived from other domains, such as biochemistry,
genomics, immunology, materials science etc. and the semantic web has significant offerings to make
when moving towards to goal of inter-disciplinary data mashups. The twin pillars of the URI and the
web of linked data can be sure to have a profound impact on the way in which science will be carried
out in the 21st century. For chemistry, this will, for example, mean that once a resource such as a
molecule or a chemical substance has been defined, it can then be linked to data about its properties,
physico-chemical characteristics etc and also to knowledge that other disciplines outside of chemistry
have about this compound with great ease and it will be possible to analyse for other information such
as co-occurence of this compound with other compounds, research activity involving this and related
compounds, citations etc..
Chemists produce and report vast amounts of data every day. The Chemical Abstracts Service (CAS)
indexes over 10,000 new substance every day.1 This data is mainly derived from typical scientific
publications. On top of this, there is an even larger amount of data which is contained in (electronic)
laboratory notebooks, reports and patents. The widespread adoption of data from high-throughput
experimentation is further swelling the data deluge. The one common characteristic, which data from
all of these sources has, is the fact that it is usually contained in documents in a completely
unstructured form, which makes it exceptionally hard to search, to retrieve and to mash up.
Given the importance of mashups then and the unstructured nature in which chemical information is
normally produced and recorded, one important task is to develop technologies that allow the
extraction and structuring of chemical data and thus the "semantification" of chemistry. Roughly
speaking, a "semantification workflow" could look like this: (a) identification of sources containing
chemical information (typically scientific papers, scientific theses, blogs, wikipedia entries, other websources) (b) identification and extraction of chemical entities and other chemistry relevant information
and (c) markup of the extracted entities /data in XML or RDF. The identification and extraction process
is the important step here and natural language processing (NLP) technologies and parts of speech
taggers (POS) are the tools of choice. Although there is considerable interest for this type of workflow
in other domains such as the biosiences and medicine and a number of tools have been developed by
both commercial (e.g. Temis, Linguamatics etc.) and academic groups (e.g. GENIA, PennBioIE etc.),
chemistry lags sadly behind, although a number of reports concerning the extraction of chemical
entities from the literature have been reported over the last several years.2-5
2. Extraction of Chemical Information from unstructured Text
The prime open tool for the extraction of chemical entities is the OSCAR 3 system 6 and we will show
how entity extraction can be accomplished using OSCAR 3 and a part-of-speech tagger provided by
NLTK provided by the Natural Language Toolkit (NLTK).7 OSCAR (Open Source Chemistry Analysis
Routines) is an open source application and part of the SciBorg project 8 for the deep parsing and
analysis of scientific texts, but can also be used in a standalone or integrated with other NLP systems.
NLTK is a suite of open source modules, data sets and tutorials supporting research and development in
natural language processing. It uses symbolic and statistical natural language processing. NLTK will be
used here to find the parts of speech and then extract the key-phrases.
To demonstrate how we apply these tools we will walk through a simple chemical synthesis procedure
taken from a typical PhD thesis8 in organic chemistry:
"nBuLi ( 1.6M solution in Et2O , 18.75 ml , 30 mmol ) was added to a stirred solution of alkyne 155 (
5.0 g , 27 mmol ) in Et2O ( 20 ml ) at -78°C and the mixture stirred for 1 h . Freshly cracked
paraformaldehyde ( mp =163-165°C ) was bubbled through the reaction mixture , which was under a
constant argon flow . After 20 - 30 min , the mixture was diluted with Et2O ( 200 ml ) and poured onto
saturated NaCl solution ( 150 ml ) , the phases were separated , and the aqueous layer extracted with
Et2O ( 2 x 50 ml ) . The combined organic phases were dried ( MgSO4 ) , filtered and concentrated in
vacuo . Purification by flash column chromatography ( eluent PE : Et2O 4:1 to 1:1 , gradient ) yielded
alcohol 156 ( 4.67 g , 21 mmol , 81 % ) as an oil."
Figure 1: An example of a chemical synthesis procedure from a PhD thesis.
So far the only structure we have in this text is the title of the preparation and its content. We use
OSCAR to identify the chemical names in this text :
"nBuLi ( 1.6M solution in Et2O , 18.75 ml , 30 mmol ) was added to a stirred solution of alkyne 155 (
5.0 g , 27 mmol ) in Et2O ( 20 ml ) at -78°C and the mixture stirred for 1 h . Freshly cracked
paraformaldehyde ( mp =163-165°C ) was bubbled through the reaction mixture , which was under a
constant argon flow . After 20 - 30 min , the mixture was diluted with Et2O ( 200 ml ) and poured onto
saturated NaCl solution ( 150 ml ) , the phases were separated , and the aqueous layer extracted with
Et2O ( 2 x 50 ml ) . The combined organic phases were dried ( MgSO4 ) , filtered and concentrated in
vacuo . Purification by flash column chromatography ( eluent PE : Et2O 4:1 to 1:1 , gradient ) yielded
alcohol 156 ( 4.67 g , 21 mmol , 81 % ) as an oil."
Figure 2: An example of a chemical synthesis procedure from a PhD thesis after markup using the
OSCAR 3 system.
OSCAR marks up chemical entities contained in this paragraph using a mixture of SciXML and a
technology developed by our group here in Cambridge. The first part of the figure 3 (A) presents the
title of another paper and the first sentence of the abstract in natural language. Part (B) shows the
same sentence after markup through the OSCAR 3. In this example, chemical entities such as “oleic
acid” or “magnetite” are marked up as chemical moieties (type=”CM”) and additional information,
such as in-line representations of chemical structure (SMILES and InCHI) as well as ontology terms and
other information can be added.
Figure 3: Markup of
Chemical Entities via the OSCAR 3 system.
Once the chemical entities have been marked up in this way, we can use the Natural Language
Processing Toolkit to determine the syntactical structure of the text. By doing so, we are able to
determine quantities and several types of experimental conditions. Crucially, it is also possible to
detect actions such as addition, dissolution, extraction etc. in the text (Figure 4).
Figure 4: 'Action' Phrases
marked up using NLTK.
At this stage, information has now been marked up. The resulting parse tree is stored in XML (Figure
5) .
Figure 5: Parse tree stored in XML format.
The parse tree is then converted to RDF. We will simplify at this point and imagine that the resource
www.foo.bar/preparation-1 is a unique URI :
Figure 6: Simplified RDF Graph of the synthesis procedure from figure 2.
What results is in effect an RDF-graph based representation of the above paragraph. Not only does the
graph contain all the compounds involved in the preparation (and which themselves could have other
information such as properties, supplier data etc associated with them), but also their roles (which can
be deduced from actions etc.) and other information such as yields etc... . Assignment of roles allows
the classification of compounds into reactants and products, solvents, reagents, catalysts etc. Once
chemical information is stored in this way, it now becomes feasible to search for experiments by
parameter: an example query would be to search for all experiments that have yields over 80% and
that use substances which are solvents and have boiling points below 50 C.
When this approach is applied to a whole document rather than a single paragraph, it becomes possible
to draw "chemical topic maps" from the literature. In documents detailing chemical experiments, it is
common to assign numbers to each occurring chemical entity. By tracking the compounds involved in a
synthesis procedure and the reactions they participate in, it is possible to generate "reactant yields
product" graphs (figure 7).
Figure 7: ''Reactant Yields
Product” Graph.
In the example in figure 7, the compound with ID 155 is transformed into compound 156 during a
chemical reaction. If we now plot this for all the transformations identified in a document, we arrive at
"topic map" of chemical transformations (figure 8).
Figure 8: Topic Map of a
Thesis.
This snapshot provides a concise overview over the entire document. The colours represent the colours
of the products (identified from NLP) and the shapes of the nodes represent the states of compounds
(oil = circles, solid = squares, crystal = diamond). This information can be directly parsed out of the
text or inferred from information which has been extracted. However, the shapes of the graphs and the
connectivity between compounds also holds information: were chemical syntheses performed in
parallel or did isomers (i.e. compounds which have the same chemical composition but a different
physical arrangement of atoms) form during the reaction? This and other information can potentially be
gleaned by inspecting the shape of the graph.
While the above discussion has illustrated some technical solutions to the problem of semantification of
unstructured information within the domain of chemistry, the real challenge remains one of access.
Sources of chemical information and documents are typically proprietary and closed access and content
providers such as science, technology and medicine publishers are taking active steps to prevent use of
the technologies we have described here for the extraction of scientific data from their sources. The
situation is aggravated by the fact, that chemistry, unlike other sciences, has not evolved a culture or
tradition of data sharing, which means that both the technological infrastructure as well as the
"mindshare" are currently absent.
3. Summary and Conclusions
Chemistry is a central science which is at the heart of many high-value industries. While the volume of
data produced by chemists is high, chemical information is often unstructures and thus hard to search,
retrieve and mash-up. However, structuring and thus semantification are possible using a combination
of natural language processing and parts-of-speech tagging. The results from NLP and POS processing
can be translated to RDF, which, in turn, can be used for both the visualisation of topics and
relationships in documents and the quick search, retrieval and mashup of chemical information. The
oftentimes proprietary and closed nature of chemical information sources presents a serious obstacle
to the semantification of chemistry.
Semantic Chemistry
Author(s):
Nico Adams
Executive Summary
Chemistry is an important and high-value vertical in the modern world and the "semantification" of
chemistry will be crucial for further rapid innovation not only in the discipline itself, but also in related
areas such as drug discovery, medicine and materials design. This article provides a short overview
over the current technological state of the art in semantic chemistry and also discusses some
obstancles, which have, so far, impeded the widespread uptake of chemistry in the domain.
1. Introduction
Chemistry is arguably the most central of the physical sciences and at the heart of many fundamental
industries: developments in chemical science very directly affect sectors such as the pharmaceutical
and medical industry, the producers and processors of modern materials such as polymers and, of
course, the chemical industry itself.
In modern science, it is important to realise, that most of the truly exciting scientific and technological
progress now happens at the interfaces between two or more scientific and technical disciplines. As
such, the development of new knowledge or products can, from an informatician’s point of view, be
considered to be an exercise in the integration of data from different scientific domains. Chemistry
overlaps with almost all domains of modern science, from pharmacology, biochemistry, toxicology to
genetics and materials science.
As such, it is of prime importance to develop a comprehensive semantic apparatus for the discipline,
which can contribute to the “data integration” process. This short article is subdivided into three
parts. In the first part, it will discuss the state of the art in semantic chemistry at the time of writing
(early 2009), the second part will look at current efforts in “semantification” within the domain of
chemistry and the third part will discuss some of the technical and “socio-political” obstacles semantic
chemistry is facing today.
2. The Current “Semantic Chemistry Technology Stack”
The general semantic web toolkit in common use today consists of three major components: XML
dialects, RDF(S) vocabularies and OWL ontologies (Figure 1).
Figure 1: The semantic
layer-cake.: (Copyright © 2008 World Wide Web Consortium, (Massachusetts Institute of
Technology, European Research Consortium for Informatics and Mathematics, Keio University). All
Rights Reserved. http://www.w3.org/Consortium/Legal
Let us look at each of these components in turn and how they have been applied to the field of
chemistry.
2.1 Markup Languages for Chemistry
In terms of markup languages, the foremost and most relevant markup language pertaining to the
realm of chemistry is Chemical Markup Language (CML), developed over the last decade by MurrayRust, Rzepa and others.[1-7]
CML is designed to hold a large variety of chemical information, such as molecular structures (the
spatial location of and the connectivity between the atoms that make up a molecule), materials
structures (in particular polymers) as well as spectroscopic and other analytical data and also
crystallographic and computational information. An example of the CML-based representation of the
molecular structure (i.e. the atomic composition of a molecule and the spatial arrangement and
connectivity of the atoms making up a molecule) is shown in figure 2.
Figure 2: CML document
describing the 2-dimensional molecular stucture of the styrene molecule.
The CML document describes an entity of type <molecule>. The <molecule> is a data container for two
further data containers called <atomArray> and <bondArray>. The <atomArray> element contains a list
of all the atoms present in the molecule, together with IDs, element types and, in this case, 2D
coordinates specifying the spatial arrangement of atoms in the molecule. The <bondArray> element by
analogy contains a list of bonds, bond IDs, a specification which atoms are connected by the bond and
the bond order (is it a single, double, triple or any other type of bond?). Furthermore, CML can hold
many different types of other annotations on atoms, bonds and associated chemical data. CML was
recently extended to deal with fuzzy materials such as polymers, which also introduced the notion of
introducing free variables into an otherwise purely declarative language, by injecting XSLT into
specifications of CML and evaluating expressions in a lazy manner.[7]
Scientific and chemical information in free unstructured text such as scientific papers, theses and
reports can be marked up in an analogous manner. Figure 3 shows the markup of a sentence contained
in a scientific paper using a mixture of SciXML and a technology developed by our group here in
Cambridge.
Figure 3: An abstract (ref39) (A) prior to markup, (B) after markup with OSCAR 3.
The first part of the figure (A) presents the title of the paper and the first sentence of the abstract in
and (B) shows the same sentence after (automated) markup through the OSCAR 3 natural language
processing system.[8] In this example, chemical entities such as “oleic acid” or “magnetite” are
marked up as chemical moieties (type=”CM”) and additional information, such as in-line
representations of chemical structure (SMILES and InCHI) as well as ontology terms and other
information can be added.
Other markup language of relevance to chemistry include Analytical Markup Language (AnIML),[9]
ThermoML – a markup language for thermochemical and thermophysical property data,[10] MathML[11]
(Mathematical Markup Language) and SciXML.[8, 12] Furthermore, Indian researchers have recently
reported the development of an alternative to CML for the markup of chemical reaction
information.[13, 14]
2.2 RDF Vocabularies
While the ecosystem for markup languages in chemistry is relatively well developed, the same cannot
currently be said for the availability of RDF vocabularies for the domain. The most notable efforts were
reported by Frey et al. as part of the CombeChem project.[15-17]The proposed vocabulary provides the
basic mechanism to describe both state-independent (e.g. identifiers, molecular weights etc.) and
condition-dependent (e.g. experimentally determined physical properties where the property is
dependent on, for example, measurement or environmental conditions) entities associated with
molecules, as well as provenance information for both molecules and data (Figure 4).
Figure 4: Snapshot of the
CombeChem RDF vocabulary for chemistry.(15)
Furthermore, the same authors have also modelled a synthetic chemistry experiment in RDF.[18] There
are sporadic efforts to model aspects of molecular structure in both RDF and OWL, but this must be
considered to be developing work at this stage.[19, 20] In further studies, RDF has been exploited to
the purposes of publishing in the chemical domain[21, 22] and for developing technologies which could
lead to the generation of “research interest” (social) networks for chemists.[22, 23]
While at least some RDF vocabularies for chemistry are therefore available, what is decidedly missing is
the availability of mashup examples. This can be explained by the difficulty associated with getting
hold of chemical data: unlike biology or physics, chemistry has not (yet) developed a culture of data
sharing and is extremely conservative in its adoption of a more open culture. We will discuss this
further below.
2.3 Ontologies
Ontologies are computable conceptualisations of a knowledge domain and thus crucially important for
adding "meaning" to data. To date, only few attempts have been made to construct formal ontologies
for chemistry. Very early attempts predate the arrival of the semantic web and indeed the internet: in
the 1980s, Gordon considered the the syntax, semantics and history of structural formulae as well as
the semantics and formal attributes of chemical transformations in a set of papers, which led to a
formalised language for relational chemistry.[24-26] Somewhat later, van der Vet published
construction rules for the some very fundamental chemical concepts, such as "pure substance", "phase"
and "heterogeneous system" as the basis for the development of further axiomatisations relevant to
chemistry.[27]
The currently most widely used chemical ontology is the European Bioinformatics Institute's (EBI)
"Chemical Entities of Biological Interest" (ChEBI) ontology.[28] ChEBI combines information from three
main sources, namely IntEnz,[29] COMPOUND and the Chemical Ontology (CO)[30] and contains
ontological associations which specify chemical relationships (e.g. "chloroform is a chloroalkane"),
biological roles and uses and applications of the molecules contained in the ontology. ChEBI is stored in
a relational database, but can be exported to OBO format and translated into OWL. Other ontologies
currently maintained by the EBI are REX[31] and FIX[32]. REX terms describe physicochemical
processes, whereas FIX mainly describes physicochemical measurement methods. Again, both
ontologies are available in the OBO format. There have been other attempts to model aspects of
chemistry, such as chemical structure,[19] laboratory processes[15-18] chemical reactions,[13, 14] and
polymers[33] but these are isolated and somewhat small-scale efforts. There is currently no discernible
community effort to develop a formalisation of chemical concepts.
3. "Semantification in Chemistry"
A significant amount of chemical data is currently tied up in unstructured sources such as scientific
papers, theses and patents. As such, natural language processing (NLP) of these sources is often
required to extract relevant information and data and to add metadata . While there is considerable
activity in processing text in the biological, biochemical and medical literature by both companies (e.g.
Temis, Linguamatics and others) and academic groups (e.g. GENIA, PennBioIE) chemistry is sadly
lagging behind in this area, although a number of reports have appeared in the literature over the past
several years.[34-37] The principal open tool for the extraction and semantic markup of chemical
entities at the moment is the OSCAR 3 system, which is currently being developed by Corbett and
Murray-Rust.[8] OSCAR 3 is part of the SciBorg project[38] for the deep parsing and analysis of
scientific texts, but can also be used in a standalone or integrated with other NLP systems. A typical
example of OSCAR's output has been provided in figure 2.
4. Cultural Access Barriers to Semantic Chemistry
So far, we have only discussed the technical aspects of semantic chemistry. And while the field is in
many ways still in its infancy (note the absence of a significant body of RDF vocabulary and ontologies),
this situation is currently being addressed by a number of academic groups as well as commercial
entities and it is reasonable to expect that a substantial amount of work will become available over the
next several years, The real challenges associated with semantic chemistry are not so much of a
technological nature, but rather "socio-cultural". We have already alluded to the fact that, unlike other
scientific, technical and medical fields, chemistry has not evolved a culture of data and knowledge
sharing. Rather chemistry has ceded the dissemination of data and knowledge almost entirely to
commercial entities in the form of publishing businesses. However, as is the case in mainstream
publishing, the internet is currently in the process of destroying the business model associated with
scientific publishing (publishers justifying subscriptions and revenue by organisuing manuscript
collection, peer review, editorial work, printing and distribution to subscribers of the journal issue). As
a consequence, scientific publishers are increasingly shifting their value proposition to content, i.e.
scientific data and seem to attempt to prevent the automatic extraction of data (i.e. noncopyrightable facts) from their journals. For obvious reasons, disciplines which have already evolved
both the technological as well as cultural mechanisms for data sharing are less severely impacted by
this than chemistry, which currently has neither the technological nor indeed the cultural wherewithal
for data sharing. Sooner or later, this will adversely affect the progress of science as a whole - the
biosciences, for example, are crucially dependent on chemical data and without the ability to mash up
data from both sources, progress in biology etc. will undoubtedly be impeded.
The crucial task for anyone interested in the use of semantics in the chemical domain, therefore, is to
not only develop the necessary technology, but first and foremost to make a contribution towards
changing hearts and minds in the discipline and to create “data awareness” in practicing scientists,
which are not also informaticians. The Open Access movement is making slow and steady progress in
this (several very significant universities have recently adopted open access publishing mandates) and
the current generation of undergraduate and postgraduate students is keenly aware of the possibilities
and the promise of semantic technologies. Therefore, there is considerable reason for optimism that
we will see the transition from "chemistry" to "semantic chemistry" and full participation of the
discipline in the semantic web in the not too distant future.
5. Summary and Conclusions
Chemistry is a conservative discipline which is nevertheless staring to participate in the semantic web.
There is a considerable and useful infrastructure of markup languages available for the dissemination
and exchange of chemical data. While not currently highly developed, some first drafts of RDF
vocabularies and ontologies are also coming on-stream and good progress in the extraction of chemical
entities from unstructured sources is also being made. The main obstacle that is currently holding up
both the further development of semantics in the chemical domain and its further adoption as a
technology is socio-cultural in nature: to date, chemistry has not evolved a culture of data sharing and
therefore neither the cultural nor the technical mechanisms are in place, which results in a scarcity of
available data sets. Nevertheless, the increasing adoption of open access and the further penetration
of semantic technology into chemistry will force change to occur and there is every reason to remain
optimistic.
Deploying Semantic Technology Within an Enterprise: A Case Study of the UCB Group Project
Author(s):
Keith Hawker
Executive Summary
This case study explains the development and deployment of the Immunisation Explorer, a newly
created business application within UCB Group that has been developed to exploit the semantic
services provided through the Metatomix Semantic Platform.
Introduction
This application brings data together from a varied range of systems into a consolidated view as new
antibodies are registered, allowing the scientists to start to answer critical questions, such as;
“What immunisation regime produced this antibody?”
Through the deployment of this application, UCB have been able to rapidly integrate data from many
different sources ranging from spreadsheets to large Oracle databases to help their business users
address dual objectives:
Targeting Research – By creating a thorough life-cycle view of an antibody at the point of
antibody registration as well as showing all related information correctly through primary
testing, secondary testing, immunization regime and back to the conditions of the animal
Utilizing Resources – By creating a scheduling-based view showing who is working on the
project, when tasks are scheduled for completion and when tasks are completed
What started as a tentative step to explore the capabilities of semantic technology has now blossomed
into a giant leap by helping users make informed decisions.
UCB is a global biopharmaceutical company based in Belgium, with operations in more than 40
countries and revenues of €3.6 billion in 2007. The company is a recognized leader in treatments for
allergy and epilepsy, and in the rapidly emerging field of antibody research, particularly in conjunction
with proprietary chemistry.
Within all biopharmaceutical companies, the cost and effort invested in new entity discovery, both
chemical and biological entities, is immense. Targeting research into the most productive areas and
effectively utilizing available resources are two key objectives for any company in the
biopharmaceutical market. The problem isn’t a lack of data, but rather an overload of raw data,
spread across entirely different IT systems, with no easy way of understanding it as a whole.
Through the deployment of the Metatomix Semantic Platform, UCB is able to rapidly integrate data
from a rich discovery process achieved through a combination of semantic modelling, non-invasive data
gathering from existing data sources and rule-driven business process-led behavior. A central capability
of the Metatomix platform is the enactment of policy-based behavior that responds to what is known,
at any point throughout the query. The policy engine is configured to know what data sources are
available and is able to trigger the appropriate query, receive data from that data source, transform it
into resource description framework (RDF) and make it available to the case for assessment.
This architecture is illustrated in the diagram below:
This process enables many different sources to provide a consolidated view of all relevant information,
enabling UCB to address dual objectives:
Targeting Research – By creating a thorough life-cycle view of an antibody at the point of
antibody registration as well as showing all related information correctly through primary
testing, secondary testing, immunization regime and back to the conditions of the animal
Utilizing Resources – By creating a scheduling-based view showing who is working on the
project, when tasks are scheduled for completion and when tasks are completed
What started as a tentative step to explore the capabilities of semantic technology has now blossomed
into a giant leap by helping users make informed decisions.
The Project
The starting point for the use case was the registration of a new antibody following its sequencing. At
this point, the scientists want to be able to view all related information and have many different
questions asked. One of the most important questions being:
“What immunization regime produced this antibody?”
However, at the point of registering a new antibody, very little is known. It isn’t possible to raise
queries against multiple data sources about an antibody, as not enough information is known to be able
to furnish the queries. A knowledgeable user could traverse the different systems by connecting the
dots, but this is hugely time-consuming, even if the user has been given comprehensive data access.
The Immunization Explorer
The business use case developed within UCB takes advantage of the enrichment framework, and
defines what has been called the “Immunization Explorer” application. This application is the first
point along the entire antibody research life-cycle, and it will extend this application to embrace many
other similar entry points where scientists can look across all the relevant data.
The Immunization Explorer starts with the registration of a new antibody. The application creates a
case and proceeds to collect all the relevant information associated with this case by enacting a
number of iterative queries through the different data sources. This information is able to trace back
through secondary testing and primary testing to the immunization regime that initiated the project.
This is known as the antibody enrichment cycle.
This cycle constructs a consolidated view of all relevant information associated with any newly
registered antibody through the different phases of the project. In this way, scientists are able to
evaluate what immunization regimes are leading to the production of antibodies, with and without the
right properties. Scientists are able to track which sample is the source of the new antibody, and
identify the culture plate and associated assay plates which contain samples from the same source.
This is an example of semantic model-based integration working in conjunction with a process-centric
rules engine to create an application that can respond to the level of knowledge that is known at any
point and drive enrichments based on this level of knowledge. This enrichment cycle navigates through
and collects data from a wide range of systems, transforming the data into RDF and assembling it into a
single data model within the Metatomix Semantic Platform. The resulting model is then available to be
analyzed in many different ways by the user. This is illustrated below:
Triggering the Antibody Enrichment Cycle
The antibody enrichment cycle is triggered in one of two ways. The first method is an automated backend process that passes the list of candidate antibodies through to the Metatomix Semantic Platform.
This technology responds by pre-preparing the information for a user through the creation of individual
cases for each antibody. The information is also enriched for each user so it is ready to be queried
through the User Interface.
The second method allows a user to enter queries directly through the User Interface. In this scenario,
the user input triggers the antibody enrichment cycle, causing a case to be created and for this to
trigger the call-out to the different data sources, the conversion of the different data into RDF and the
presentation of the consolidated information back to the scientist in the User Interface.
Creating the Ontological Model
A range of ontologies have been developed to support the data integration requirements within the
antibody research area which provide a common model across both new biological and chemical
entities.
The concepts defined in these ontologies cover the concepts relating to the data surrounding
experiments, tests, test results, and the immunization regime. As well as, the project life-cycle
concepts relating to stages of the project and people aspects, such as who is working on the project
and their reporting structure.
Collectively, these ontologies create a single conceptual model within which all the disparate data can
be understood within a common framework, both to allow scientists to look across all relevant data
with each experiment and to allow project managers to stay current on each projects progress.
A subset of these concepts and their relationships are illustrated below:
Bringing the Data Together
Following the creation of the ontological model, each data source is mapped to ontologies so data can
be collected, transformed and inserted as instance data that is understood within the common model.
In this way, it becomes immaterial as to which data source is the source for any particular piece of
information as all data can be seen, accessed and interpreted in a single consolidated view.
For each data source a process chain is constructed using a library of pre-built utilities supplied as part
of the Metatomix Semantic Platform that provide connectivity and data transformation methods. This
significantly increases the speed with which data integration can be achieved.
Acting on the Data
The Policy Engine, provided with the Metatomix Semantic Platform, provides a wizard-based method
for constructing rules that can assess a level of knowledge at any point and can configure necessary
actions to be taken based on this knowledge.
Policies are constructed in order to control a set of service requests that invoke specific data queries,
the collection of data from different data sources and the transformation of data into RDF within the
common model.
A Single Application with Different Uses
As described above, the Immunization Explorer provides a consolidated view of all information relating
to an antibody, collected as a result of its registration.
At the same time as this information is being assembled, further data queries are made into a range of
other systems that explore which users are working on a project, and the projects status. Determining
the status is often through interpolation across data sources and inferring the stage a project has
reached. For example, detecting that a proposed project does not yet have a start date can be
interpreted with the status “awaiting ordering of animals.”
This information is collected, interpreted and presented in the Immunization Scheduler Interface,
which is used by project managers, rather than scientists.
Conclusion
UCB began with the idea that using semantic technology could help solve the problem of efficiently and
cost effectively bringing together large amounts of raw data. With the help of Metatomix Semantic
Technology, the Immunization Explorer project was completed within two months and is going into
production.
There has been great enthusiasm engendered within the business community to extend semantic
technology similar to that used with the UCB/Metatomix project across other enterprises. Semantic
technology has proven to effectively bring disparate data together within an enterprise and continued
success stories like the UCB/Metatomix project further show the strong potential this technology
possesses.