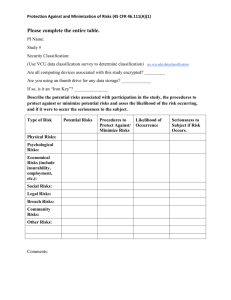
Annotated bibliography

Comparative Genomics Projects of Bacillus Phages
Spring 2015 Phage Lab
For the rest of the semester, we will work on small group projects to gain a better understanding of similarities and differences in the sequenced Bacillus phages discovered by VCU Phage Hunters and others. The research questions you explore are almost always based in biology, but use a bioinformatics approach for investigation. Your data will be included on a poster for presentation at the VCU undergraduate research festival, presented at the 7 th annual SEA PHAGES symposium at HHMI’s Janelia
Farms Research Campus in June 2015, and described in a final paper you submit to me to complete your course requirements.
Note: If you are successful in your project and produce an interesting piece of analysis, we will include your data in a paper submitted to a research journal! So, dream big- your ultimate goal as part of this scientific process should be to produce one publication quality data figure or table for a paper describing and interesting feature or characteristic of VCU’s Bacillus phages.
You can work alone, or in small groups. You are welcome to pick your own project or to try one that I suggest. You must use one of our VCU phage sequences!
Important Dates:
March 4 th , 16 th : Discuss draft of project plan, with four supporting literature sources identified. Be prepared to verbally discuss why each article is relevant to your project.
March 18 th : Submit project plan and annotated bibliography (can be completed by group).
April 22 nd : Present poster at VCU undergraduate research festival
April 27 th : Last class. Final symposium sometime that week.
May 8 th : Final paper is due.
Some project suggestions are:
1.
Examination of promoter regions and determination of consensus -10/35 sequences. Can you derive different consensus promoters that regulate early/middle/late gene expression?
2.
Comparison of bacillus phage sigma factors with sigma 70 from host or characterized sigma factors from other phage. Can you determine the role of Phrodo or other VCU phage sigma factors?
3.
Determination of consensus SD sequence. We think the Nigalana consensus is similar to E coli
SD sequence used by DNA Master. Is this true for other Bacillus phages?
4.
Mosicacism/recombination in bacillus phages- by GC content profile, by protein content, by other? See examples in my slides.
5.
Other items of interest- restriction endonuclease? Genes of bacterial origin? You are welcome to example a protein family and/or try to predict function of proteins without a prediction.
6.
Complete annotation of other phages- if you choose this, you must help determine genome ends and complete a small comparative genomics project (generate summary data table, dotplot comparision, identification of potential recombination event, single gene family phylogeny, etc.).
Where to start? The Literature: Each person/group should start their project by reading the literature.
Don’t just jump in without knowing what to look for, and what is known! I encourage you to read to find a figure demonstrating a bioinformatics approach that you want to replicate with our group of
phages. I’ve posted an article on blackboard supporting several of the projects listed above and you can also take a look at the slides showing work of past students.
The best way to access the literature (for me) is through the PubMed link under the databases dropdown on the VCU library homepage. http://www.library.vcu.edu/ Beware, sometimes students have trouble retrieving subscriptions through the VCU Portal link to the library. If you are off campus, you should access the library directly and sign in with your eID for access to online journals.
Develop a Research Question: On March 4 th /16 th , I will meet with each project group to talk about your research question. Envision your goal and how you will get there. Sketch out what you think your final data will look like. We will also troubleshoot your literature search supporting your topic, and make sure you understand the example I have provided. You must have identified your 4 additional literature sources before this meeting.
Annotated bibliography- Due March 18 th , develop an annotated bibliography describing specific data/analysis from 5 papers that you can use to support your project. It is absolutely o.k. to share and discuss papers in your group, and to write this bibliography together. Your bibliography could include the publication describing the tool you will use (yes, there is a paper describing BLAST, for example), and several primary research articles examining your topic in other phages. You should document the reference information and relevant data/figure in a shared wiki post or word doc, and write a mature paragraph describing that study’s input data, their analysis tools/approach, their findings, and what the data tell you about your approach or your phage genome component. This info will go into the discussion of your final paper as ‘evidence’ and ‘reasoning’!- so put some significant effort here and it will save you work in compiling your final paper.
Data: Pulling together the data you need is generally a bit of a quilting project- combining data from several sources. Genbank: many phage genomes and their annotations are available there. Blackboard:
All sequenced VCU phage genomes and their autoannotations will be posted on blackboard. Only
Nigalana is annotated and in Phamerator. Phamerator: a powerful tool and good source of gene sequence data for any phage that is included in the database. Ask me for anything else you need!
Ongoing: I will meet with each project group to review your project plan, including the question you will attempt to ask, the data and tools you will use, your general approach. You should be prepared to update the class on your project in a lab-meeting style forum. Be prepared to answer questions from me and the group- verbally, on the dry erase board, etc.- and to show work-in-progress.