Activity Protein Structure
advertisement
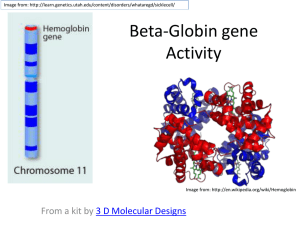
Name: _________________________________________ Date: __________________ Block: _________ Tutorial: The use of databases has been an invaluable tool for biological researchers in many fields such as protein research. The main resource for most biochemists is the Protein Data Bank (PDB). Biochemical researchers worldwide involved in protein structure determination submit their structures to this database along with their publication in the scientific literature. The PDB organizes this information to make it easy to search and also to display the structures. Anyone with access to the internet can view the structure of any protein of interest found in the database. Another valuable resource can be found at the National Center for Biotechnology Information (NCBI). This site provides links to a diverse set of databases along with many links to software that allows data to be viewed easily in a meaningful way. This activity will focus on software linked to the “Structure” section of the database. You will need to install the free viewer “Cn3D” by going to http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml and following the directions. Activity: Go to the following site: http://www.ncbi.nlm.nih.gov/Structure/MMDB/mmdb.shtml. From the drop down menu select “structure” and follow the directions below. Below is a screen shot. You can search for structure based on name or if you already know the official number assigned for the structure, you may enter that. We are going to first look at a particular structure of human hemoglobin as an example, but there are many different hemoglobin structures available. Try a search of the word “hemoglobin” and see what I mean. Type the word hemoglobin in the box next to the word “for” at the top of the screen. 1 Question #1: How many entries are listed for hemoglobin? (This is written in blue on the “all” tab.) We want to look at the particular hemoglobin structure (human hemoglobin) with the PDB number 2H35. Type 2H35 in the space where you previously typed hemoglobin. That should bring up a screen with a small structure picture of 2H35 (human hemoglobin) on the left. Click on the structure picture. That should bring up a page with a variety of information that looks like the screen shot below. There is a reference to the publication, a link to the abstract, a link directly to the PDB (click the 2H35 on the top by “PDB” if you want to see that), and other information. Most importantly however, this is how we can open the structure viewer. Click the “View Structure” button to the right of the center picture as shown in the diagram above. The advantage of this software is that it provides very easy integration of the sequence information and the picture of the structure. Three windows should open on your screen, one with the picture, one with the amino acid sequence, and one with a Cn3D message log. You have several available functions here: 1) You can grab any part of the molecule and rotate it by using the left mouse button. 2) You can “zoom” the structure by using the zoom option in the view menu, or simply by keying “z” zoom in or “x” zoom out 3) You can re-center the structure by holding down “shift” while dragging with the mouse. 4) You can get it back to the original size by choosing the “restore” option in the view menu. Now let’s explore different ways of representing the structure. These are changed by use of the “style” pull down menu at the top. Open that, then click “rendering shortcuts”. This gives you five different style options. For each of the style options you can also make the side chains visible or invisible by using the “toggle side chains” button at the bottom of the “rendering shortcuts” menu. Try all the style options both with and without the side chains. Question #2: Which do you think is the easiest way to view the protein model? 2 At any time, if you wish to save a picture of the structure as you currently have it displayed, you may create a PNG file by using the file menu and “export PNG”. Create a PNG file of hemoglobin in the worm style with no side chains showing. View the next two proteins for comparison with hemoglobin. Create similar PNG files for these two proteins as well: Src Kinase (3DQX) and Cytochrome C (3CP5) Once you have your PNG files then access http://www.rcsb.org/pdb/home/home.do and use this site to find out additional information about the proteins that you have just explored. Finally, complete the following questions using your images as well as the protein data bank. Assignment: 1. Import all three PNG into this word document. Please align then side by side with captions showcasing which protein is which. 2. Describe the various levels of protein structure: primary, secondary, tertiary, and quaternary that you are able to see in each protein. (NOTE: Tertiary may be a bit difficult so use your best judgment as to what is possibly is occurring at this level --- You can do this in your captions if you choose.) 3. Create a table that compares the three proteins in terms of similarities and differences in secondary structure and quaternary structure. 4. Within your table include a column or row that describes what the protein is and what its various functions are. Use the protein data bank to figure out this knowledge. 5. When complete, please upload your document to GoogleDocs and share with Mrs. Flick. 3