Separating the Effects of Genetic Drift and Natural
advertisement

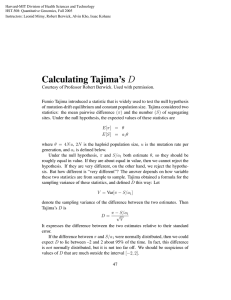
Mathbio proposal: Separating the Effects of Genetic Drift and Natural Selection using a Modification of Tajima's D statistic Mentors: Tony Weisstein and Pam Ryan Project Description: Several statistical measures have been devised to infer past evolutionary forces from current patterns of genetic diversity in sequence data. Tajima's D detects increases or decreases in overall genetic diversity, but cannot assess the relative contribution of genetic drift and natural selection to the variation observed. We previously wrote a computer simulation which starts with a population of 500 identical DNA sequences and allows the population to randomly mutate and recombine for 20,000 generations. We then select a sample of 20 organisms (from the last generation) of our population and classify the mutations in the sample as synonymous or nonsynonymous. Finally, we calculate Tajima's D separately on each set. In principle, Tajima’s D applied to nonsynoymous mutations (Dnon) should measure the effect of both drift and selection on the population, while Tajima’s D applied only to synonymous mutations (Dsyn) should measure the effects of drift within a population. Subtracting these quantities, we can measure the effects of selection (Dnon – Dsyn). In theory, this refinement to Tajima’s D will allow us to determine not only whether diversity is increasing or decreasing, but also whether the increase or decrease is due to a bottleneck or purifying selection (in the case of decreasing diversity) or subdivision or diversifying selection (in the case of increasing diversity). Our results to date have been encouraging; however, this method has limited power to distinguish among different scenarios of selection and drift. We propose to extend our analysis in two key ways: 1) Attempt to improve our resolution of different evolutionary models by using techniques such as Bayesian analysis, neutral networks, and support vector machines; 2) Assess the accuracy with which our method classifies mutations as synonymous vs. nonsynonymous mutations, using our computer simulation’s built-in tracker function. Skills needed: Both students will need to - be willing to get out of their “comfort zone” and learn material outside of their disciplines. - have the ability to do independent research and make hypotheses based on data. - be willing to work on an interdisciplinary team and consider other points of view. The ideal Biology student will have - at least STAT 190 or the equivalent - a background in molecular genetics and population genetics - knowledge of (or at least comfortable with) quantitative approaches The ideal Mathematics student will have - a strong interest in biology; equivalent of BIOL 107 background - some programming experience (C++ preferred) - linear algebra or higher - a strong interest in and background in statistics; especially in classification techniques References: Rand DM and Kann LM (1996). Excess amino acid polymorphism in mitochondrial DNA: Contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 13: 735-748. Williamson SH et al. (2005). Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Nat. Acad. Sci. USA 102: 7882-7887. Nei M and Gojobori T (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418-426. Tajima, F. (1989). Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics, v. 123, 585-595. Weisstein et al. (2009) Separating the Effects of Genetic Drift and Natural Selection Using a Modification of Tajima's D Statistic, Techical Report.