ESSMod Workshop Notes To Do: David: get datasets from Dave
advertisement

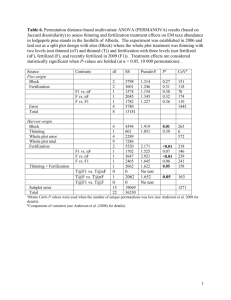
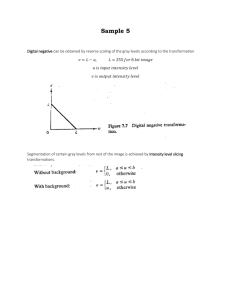
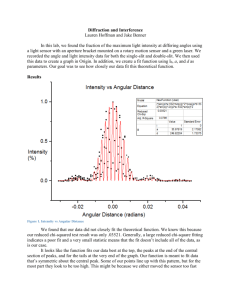
ESSMod Workshop Notes To Do: 1. David: get datasets from Dave Miller 2. David: ask for weed data 3. David: to ask Louise to create Distance-to-iDistance data conversion function 4. Janine to send her point process slides to David 5. Feb meeting agenda item: Construct schedule for getting slides and computing done in good time. Who workshop is aimed at Prerequisites: Be familiar with Distance Sampling (at a level of someone who has done the Introductory Distance Sampling workshop) Be familiar with R (give pointers to places they can learn R) Expected audience: Quantitative ecologists with interests in Distance Sampling and/or INLA A few statisticians with interests in Distance Sampling and/or INLA Schedule of presentations & Exercises Day 1 0. Introduction & Admin stuff 1. Spatial Point processes: 1.1. Describe and illustrate NHPPs 1.1.1. Intensity(=density) and log-intensity, with covariates (and splines/smooths?); difference from geostatistical process 1.1.2. Marked point processes (no longer need this for dealing with groups) 1.1.3. Likelihood function 1.1.4. Plots to illustrate 1.2. Thinned Spatial Point processes 1.2.1. Thinning NHPP with detection probability function (perhaps initially also example of thinning where thinning function is known?) 1.2.2. Effect of thinning on intensity and log-intensity 1.2.3. Likelihood function 1.2.4. Plots to illustrate Morning coffee around this time 1.3. Show how this relates to conventional distance sampling 1.3.1. With constant intensity in covered region (thinned homogeneous Poisson model) 1.3.2. With variable intensity (count method: thinned nonhomogeneous Poisson count in each segment) 2. Estimation using thinned NHPP 2.1. Bayesian inference (v. short summary relevant to above, including MCMC vs INLA) 2.2. Introduce basics of iDistance (minimal detail, including how to specify detection functions, priors; just enough to do the exercise below) 2.3. Do exercise using pre-prepared data (Johnson et al, weeds? Provide their paper) with iDistance. 2.3.1. Provide data object, provide mesh, fix priors 2.3.2. Provide R code to fit model, plot & diagnostics 2.3.3. Participants try different detection functions, different covariates, (& various smooths? These would be smooths of covariates?); select model; interpret output (including diagnostics); plot relevant stuff. Lunch around this time 2.4. PERHAPS (or as an optional extra): Do same analysis in Distance? Probably rather as an optional extra. The solutions to the exercises will be good point where we can point out the differences again, in practice as well). 3. Introduce random fields & log-Gaussian Cox processes 3.1. Random field as a smoothed error term & GMRFs. 3.2. Show how enters intensity of thinned NHPP 3.3. Illustrate lGCP by set of realisations of same process, showing “hotspots” located randomly 3.4. Simulate lGCP (from some real dataset) 3.4.1. Fit to simulated data with Distance; look at mean and variance 3.4.2. Fit to simulated data with iDistance; look at mean and variance (point being Distance underestimates variance ‘cause assumes spatial independence) 3.5. Intrinsic & other GMRFs: What are the implications for users (not so much what their definitions are) 3.6. Explain difference between Distance and iDistance 3.6.1. Distance assumes indep residuals, iDistance not. 3.6.2. Distance “binning” and count process; iDistance not-binning and continuous process in space 3.6.3. lGCP fit and predict in one; Distance separate 3.6.4. Simultaneous fit of intensity surface and detection functions 3.6.4.1. Limited detection function options currently in iDistance 3.6.4.2. Including covariates in detection functions 3.6.5. Priors for lGCP parameters Afternoon coffee around this time 3.7. Exercise: Use iDistance on weeds data Day 2 4. Detection of groups, not individuals 4.1. (Finn & Fabian ...) 4.2. Exercise (maybe with pre-prepared estimates) Morning coffee around this time 5. Model selection and diagnostics 5.1. Model selection (DIC?) 5.2. Goodness-of-fit (Joyce et al. …) 5.3. Exercise … Lunch around this time 6. More complex examples 6.1. Numerical integration and mesh construction 6.1.1. Fabian’s tutorial on mesh construction, with explanation of numerical integration. Focus on what students need to know to use methods. 6.1.2. Exercise on mesh construction Afternoon coffee around this time overlapping transects (not an issue we think) 7. spatio-temporal modelling 7.1. … 7.2. Exercise (maybe need pre-prepared example)