This Word file includes - Proceedings of the Royal Society B
advertisement
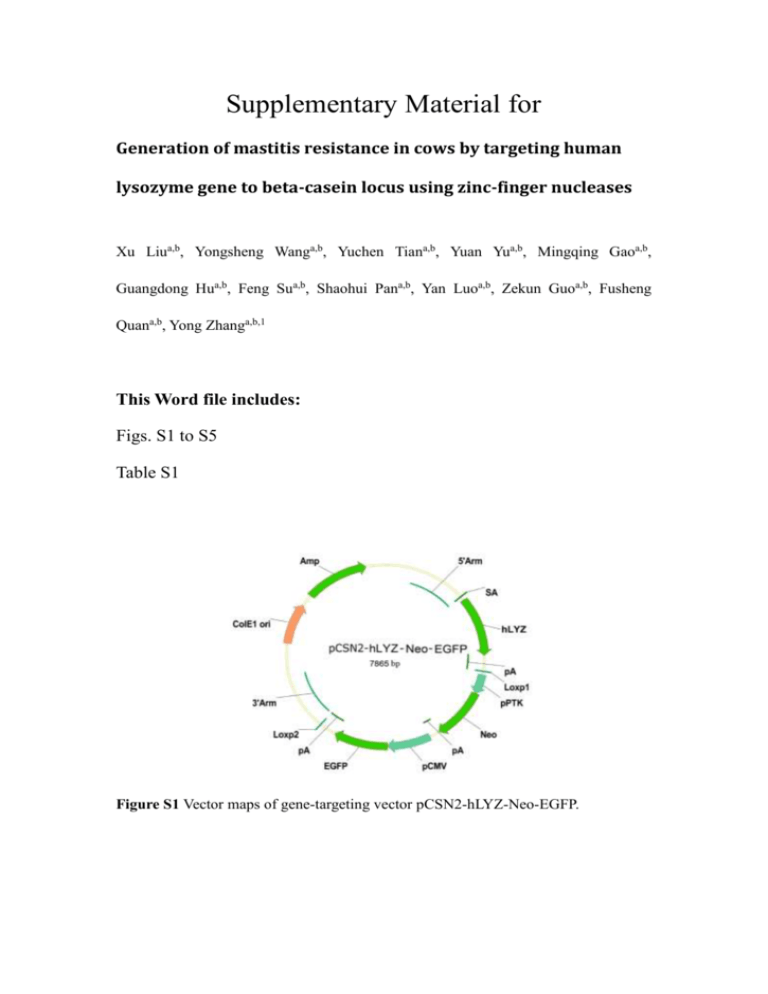
Supplementary Material for Generation of mastitis resistance in cows by targeting human lysozyme gene to beta-casein locus using zinc-finger nucleases Xu Liua,b, Yongsheng Wanga,b, Yuchen Tiana,b, Yuan Yua,b, Mingqing Gaoa,b, Guangdong Hua,b, Feng Sua,b, Shaohui Pana,b, Yan Luoa,b, Zekun Guoa,b, Fusheng Quana,b, Yong Zhanga,b,1 This Word file includes: Figs. S1 to S5 Table S1 Figure S1 Vector maps of gene-targeting vector pCSN2-hLYZ-Neo-EGFP. Figure S2 Surveyor nuclease assay of the 16 gene-targeted cell colonies at the eight most-likely off-target sites. Lane 1, DNA marker. Lane 2, non-transfected cells were used for negative control. Lanes 3–18, 16 gene-targeted cell colonies suitable for NT. Figure S3 Western blot analysis of human lysozyme production by transfected bovine mammary epithelial cells. (A) Vector map of construct used for expressing cytosolically directed human lysozyme and EGFP fusion protein. (B) Vector map of construct used for expressing secreted human lysozyme and EGFP fusion protein. (C) Vector map of construct used for targeting CSN2 locus in bovine mammary epithelial cells with ZFNs. (D) human lysozyme and EGFP fusion protein was expressed in the cells. The bovine mammary epithelial cells were transfected by pEGFP-C-hLYZ. (E) The expressed human lysozyme and EGFP fusion protein was secreted into the extracellular. The bovine mammary epithelial cells were transfected by pEGFP-S-hLYZ. (F) Human lysozyme and EGFP fusion protein expression was observed 2 days after treatment with inductive medium. The bovine mammary epithelial cells were transfected by pEGFPI-hLYZ and pZFN1 plus pZFN2. D’, E’ and F’ are the bright field view of D, E and F, respectively. Proteins in cell extracts (D) or medium (E, F) were probed with antilysozyme antibody, anti-GFP antibody and anti-casein antibody. (G, H, I) Western blot analysis for the expression of GFP, hLYZ and casein in transfected bovine mammary epithelial cells. Lane 1, non-transfected cells were used for negative control. Lanes 2– 4, Cells were transfected with plasmids containing Cyto-hLYZ-EGFP (lane 2), Sec-LysEGFP (lane 3), or Ind-Lys-EGFP (lane 4). Lane 5, standard substances of GFP, Lys and casein were used for positive control. Figure S4 Total proteins from transgenic cows hLYZ1-5 and non-transgenic cows (WT) were separated on a 15% polyacrylamide gel and visualized by Coomassie blue staining. Figure S5 Response of transgenic (n = 5) and non-transgenic (n = 5) cows to intramammary infusion. (A) Lipopolysaccharide (LPS)-binding protein concentration in serum. (B) Serum amyloid A circulating concentrations. Table S1: Theoretical off-target hotspots based on sequence identities to the CSN2 ZFN binding site Identity to target site bp ZFN monomer % 16 16 15 15 16 16 15 15 Left-F Left-F Left-F Left-R Right-F Right-F Right-F Right-R 88.9% 88.9% 83.3% 83.3% 100% 100% 93.8% 93.8% RefSeq In/Ex Chr Coordinate ZFN Binding Site DOCK11 In In In In In In In In X 24 24 6 14 14 27 18 1054469 12265288 5760409 3094503 2556274 3221824 4415251 6299052 CATCCACTATCTCAGT TCATCCACgATCTCAGT TTCATCCACTtTCTCA TTCAaCCACTATCTCA CCTATGGGACCACAAG CCTATGGGACCACAAG CTATGGGACCACAAG CCTATGGGACCACAA SKOR2 PTPRM PDGFRA XKR9 LACTB2 ZNF385D N4BP1