jam12553-sup-0001-TableS1-S2-FigS1
advertisement

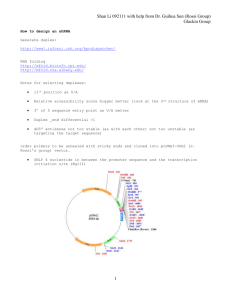
Supplementary Material Table S1 Plasmids Name Properties Reference pJOE4786.1 pBR322 pVWEx1 pJeM1 pUC8:16 pUC57hasA pJOE7706.1 pJH122.1 pJH174.1 pJH181.3 pJH182.1 pJH183.2 pJH195.2 pJH196.2 pJH197.1 ApR, lacPOZ‘ ApR, TcR, rop, rep (pMB1) KmR, rep. Ptac, lacIq KmR, rhaPBAD, rhaRS, mob, His-tag-eGFP ApR, lacPOZ, uvrA‘, vgb ApR, rep, lacZ::hasA KmR, rop, rep, Ptac lacIq, rrnB, eGFP pJeM1; ΔeGFP, hasA pJOE7706.1; ΔeGFP, hasA pJH174.1; hasC pJH174.1; vgb pJH181.3; hasB pJH174.1; glmU pJH181.3; glmU pJH195.2; hasC (Jeske and Altenbuchner 2010) (Bolivar et al. 1977) (Peters-Wendisch et al. 2001) (Jeske and Altenbuchner 2010) (Liu et al. 1994) this work this work this work this work this work this work this work this work this work this work Table S2 Oligonucleotides used for the construction of hyaluronic acid production plasmids Name S8137 S8450 S8140 S8141 S8142 S8143 S8506 S8507 S8614 S8615 Sequence (5'→3') AAAAAAACTAGTGAACAATTCTTAAGAAGGAGA AAAAAATGTACAGCTAGCTTACAGCAGTTTTTTACGGG AAAAAAACTAGTCACAGGCCTAGCCTTTTTG AAAAAAAAGCTTTCTAGATCAGTAAGCCTTGCCAGTC AAAAAAACTAGTTCAGCTTTCCTATCAAGGAG AAAAAATCTAGATCAAGCTGGCGCAATCTTG AAAAAAACTAGTATGTTAGACCAGCAAACCAT AAAAAAGCTAGCTTATTCAACCGCTTGAGCG AAAAAAACTAGTTCACTGACCGCCCAGGAT AAAAAAGCTAGCTTAGCTCTTCTTGATCTTCTCC restriction site(s) amplified gene(s) SpeI BsrGI, NheI SpeI HindIII, XbaI SpeI XbaI SpeI NheI SpeI NheI hasA hasA hasC hasC hasB hasB vgb vgb glmU glmU (a) (b) (c) (d) (e) (f) (g) (h) (i) (j) Figure S1 Microscopy of negative stained C. glutamicum cultures or culture supernatants (48 h of cultivation). (a) wild type, MEK700 minimal medium. (b) wild type, CGXII minimal medium. (c) pJH174.1, CGXII medium, uninduced culture. (d) pJH174.1, CGXII medium, induced culture. (e) pJH174.1, CGXII medium, uninduced culture supernatant. (f) pJH174.1, CGXII medium, induced culture supernatant. (g) pJH182.1, CGXII medium, uninduced culture. (h) pJH182.1, CGXII medium, induced culture. (i) pJH182.1, CGXII medium, uninduced culture supernatant. (j) pJH182.1, CGXII medium, induced culture supernatant. CGXII culture supernatants and the MEK700 wild type culture (OD600 = 8) were not diluted before staining (5 µl supernatant or culture + 5 µl nigrosin dye). CGXII cultures were diluted with H2O to an OD600 of 1.5 prior to staining. CGXII wilt type culture was diluted to an OD600 of 8 prior to staining. References Bolivar, F., Rodriguez, R.L., Greene, P.J., Betlach, M.C., Heyneker, H.L., Boyer, H.W., Crosa, J.H. and Falkow, S. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2, 95-113. Jeske, M. and Altenbuchner, J. (2010) The Escherichia coli rhamnose promoter rhaP(BAD) is in Pseudomonas putida KT2440 independent of Crp-cAMP activation. Appl Microbiol Biotechnol 85, 1923-1933. Liu, S.C., Liu, Y.X., Webster, D.A. and Stark, B.C. (1994) Sequence of the region downstream of the Vitreoscilla hemoglobin gene: vgb is not part of a multigene operon. Appl Microbiol Biotechnol 42, 304-308. Peters-Wendisch, P.G., Schiel, B., Wendisch, V.F., Katsoulidis, E., Möckel, B., Sahm, H. and Eikmanns, B.J. (2001) Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J Mol Microbiol Biotechnol 3, 295-300.