Project
advertisement
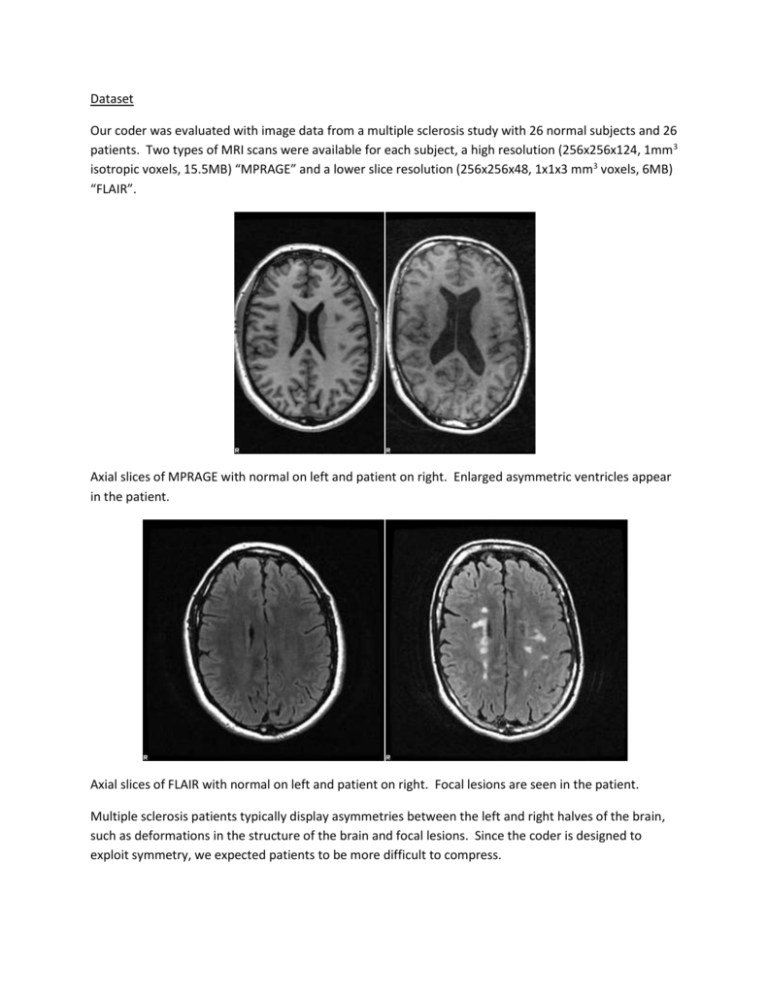
Dataset Our coder was evaluated with image data from a multiple sclerosis study with 26 normal subjects and 26 patients. Two types of MRI scans were available for each subject, a high resolution (256x256x124, 1mm3 isotropic voxels, 15.5MB) “MPRAGE” and a lower slice resolution (256x256x48, 1x1x3 mm3 voxels, 6MB) “FLAIR”. Axial slices of MPRAGE with normal on left and patient on right. Enlarged asymmetric ventricles appear in the patient. Axial slices of FLAIR with normal on left and patient on right. Focal lesions are seen in the patient. Multiple sclerosis patients typically display asymmetries between the left and right halves of the brain, such as deformations in the structure of the brain and focal lesions. Since the coder is designed to exploit symmetry, we expected patients to be more difficult to compress. Wavelet Transform A 2D wavelet transform is applied to each slice. An integer-to-integer transform is used with a lifting scheme and the biorthogonal Cohen-Daubechies-Feauveau 5/3 wavelets, as recommended by Sanchez et al. Better separation of energy could possibly be achieved with orthogonal wavelets, which should be desirable since subbands are treated independently. This is an aspect of the design that remains to be explored. The wavelet decomposition is iteratively applied 4 times on the LL subband to produce a total of 13 subbands. 4-level wavelet transform decomposition for the MPRAGE image of a normal subject. Interslice Prediction Residuals from the symmetry coding are fed into the interslice prediction. This stage starts by first chopping up the volume of residuals into chunks of 16 slices. Within each chunk of slices, the subbands are processed in raster-scan order with 16x16x16 voxel blocks. There are five prediction modes: no shift, left, right, up, and down shift by 1. On a slice by slice basis in the block, each of these shift operations is performed on the current slice and then subtracted it from the previous slice, producing a new set of interslice residuals. This process is efficiently formulated as a series of 3D convolutions. Thus, prediction is performed on a block by trying out each of the five different modes and choosing the one that produces the smallest residual MSE. These prediction modes suggested by Sanchez et al. make intuitive sense in the image domain, but it was not clear to us if they would be effective in the wavelet transform domain, since shifts there would correspond to something else quite different in the image. After plotting the distribution of chosen modes, we observed that our suspicions were correct. Interslice prediction is very rarely used when operating in the wavelet domain. It is used more often in the image domain, about 10% of the time. Results All 104 images in the dataset were compressed with our symmetry-based coder as well as generic coders including gzip, JPEG-LS, and JPEG-2000. JPEG-LS and JPEG-2000 were applied for each slice in a brain volume then compressed with gzip to reduce the overhead from any header information stored in the slice images. The following box plots show the median and range of compression ratios with each of the compression methods. Our symmetry-based coder performs on par with JPEG-LS and JPEG-2000. The generic gzip coder performs the worst because it is not specialized for images. Patients were a bit more difficult to compress than normals for all the coders. As expected, our coder has greater difficulty with the asymmetry in patient images. This is evident in the significant shortening of the upper range compared to the others. For the normal FLAIR images, the symmetry-based coder performs slightly worse than JPEG-LS/2000 but still in the same general range. There is, however, an impressive improvement for some particularly noisy patient images. The JPEG family struggles with these difficult images, doing even worse than gzip, but the symmetry-based coder handles them with aplomb. Again, patient images in general have lower compression ratios than normals. Standard Space We briefly attempted using transformations of a standard brain as an initial stage of prediction. This proved to be too limited and rate expensive in application. MR images can have varying detail and tissue contrasts but the standard brain can only effectively be used to predict another brain image if it was scanned in exactly the same way. Also, a nonlinear transformation is required to make the standard brain closely resemble the image. This requires the costly transmission of a warp transformation image that would have to be floating point accurate. Perhaps these values could be quantized with acceptable prediction inaccuracy but the added challenge of dealing with patients that appear quite different from the standard brain, which is averaged from normals, deterred us from pursuing this concept much further.