tpj12183-sup-0004-Legend
advertisement

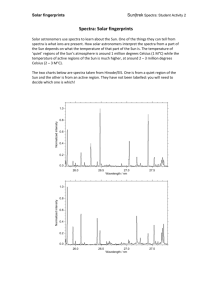
Supporting Information figure legends Figure S1. Calculation of ion intensities in selected areas of L. japonicus imprint DESI-MS images. From the images in Figure 3, the total intensities in two areas of identical size (144 pixels) from respectively the base part of the leaf (blue) and the tip (red) were calculated to provide the spectra shown. The asymmetric peak shapes in some spectra are due to averaging over several mass spectra, recorded over a longer time span during which a small drift was observed in the mass calibration. Calculations were done for m/z 104 = γ-aminobutyric acid, m/z 286 = linamarin, m/z 298 = rhodiocyanosides, m/z 300 = lotaustralin and ribesuvanins. The blue and red numbers in the spectra are the calculated cumulative intensities (ion counts) of the respective areas. The software Data Cube Explorer (AMOLF, Amsterdam, The Netherlands) was used for the calculations. Figure S2. Calculation of ion intensities in wounded and non-wounded areas of L. japonicus leaves. From the images in figure 4, the total intensities of m/z 298 (rhodiocyanosides) and m/z 300 (lotaustralin and ribesuvanins) in 36 pixels from respectively the inner, undamaged part of the ring (blue) and the damaged part of the ring (red) were calculated to provide the spectra shown. The asymmetric peak shapes are due to averaging over several mass spectra, recorded over a longer time span during which a small drift was observed in the mass calibration. The blue and red numbers in the spectra are the calculated cumulative intensities (ion counts) of the respective areas. The software Data Cube Explorer (AMOLF, Amsterdam, The Netherlands) was used for the calculations.