Sequence search
advertisement

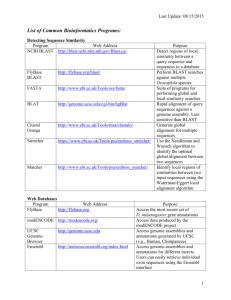
2015-12-10 Sequence search This exercise covers a general introduction to sequence search tools- BLAST and FASTA. We cannot cover all variants of BLAST and FASTA. So, we try some variants of FASTA and BLAST. We use a protein sequence from C. elegans for this exercise. You can find the protein sequence at: http://structure.bmc.lu.se/courseExercise/introBioinf.html (or at http://bit.ly/1A7HLc2). FASTA GO to FASTA website (http://www.ebi.ac.uk/Tools/sss/). Choose protein FASTA and then a protein database. Copy the sequence from the link above and paste in the appropriate text box in the form. Click on “More Options” to see more options. Check the default values for the parameters. You can set Annotation Features to Yes. But, you can also choose to see and hide the annotations from the results page. Submit the job. On the results page, you can see a table of the similar sequences. You can change the view of the results page. There are different tabs below the EMBL-EBI main menu. You can choose other views. On the left side, you can choose to show/hide annotations and alignments for the selected sequences. 1. Find the name of the sequence in UniProtKB. Find a homolog in human and report its accession and E-value. 2. Use a protein structure database and perform FASTA search. Report the PDB ID and Evalue for the best hit. Also find one more PDB ID and its E-value in the results. Do not close the FASTA search results and proceed with the BLAST search. BLAST GO to NCBI BLAST website (http://blast.ncbi.nlm.nih.gov/ ) and choose protein blast to search a protein sequence against a protein database. First check the default parameters. What is the default matrix? Are the default matrices in BLAST and FASTA the same? Use the given sequence to blast search using following parameters Database: Non-redundant protein sequences (nr) Algorithm: blastp Matrix: BLOSUM62 1 2015-12-10 Max target sequences: 1000 If you did not find some parameters, click on “Algorithm parameters” to find more parameters. 1. Are there any domains detected in the sequence? Which domains are present in the sequence? (Click on the graphics to see more about domains) 2. Are there any sequences that match exactly (100% identity) with the query sequence? a. What is the score and E-value for the exact match? Is it significant? In the Descriptions section, you can click on the Description to see the alignment between query sequence and the hit. To learn more about the protein, click on the Accession on the last column of the table. 3. Are there similar sequences in other organisms as well? Find five other organisms in your result except C. elegans. 4. What is the score and E-value of the last hit? Do you think this sequence is significantly similar? Why? Open a new tab and find ‘Smart BLAST’ from blast homepage. It is something new in BLAST. Try it out and compare the outputs of normal BLAST and SMART BLAST. Once you finish, you can close SMART BLAST results but DO NOT close normal BLAST results. Now open a new tab and perform another blast search using the same sequence and following parameters. Organism: Chicken 5. Find the best hit in chicken in your previous blast search and the last blast search. Compare the two sequences and their E-values. a. Are the two sequences same? i. If no, why do you think you have two different best hits from the same organism? b. Do they have the same E-values? Should they be the same or different? Why? Now, find similar sequences that have 3-dimensional structure in PDB. 6. Run another BLAST using the database Protein Data Bank proteins (pdb). And make sure to keep the organism field blank. 7. Does the query sequence have a 3-dimensional structure? What is the Accession? 8. Do you think the first hit is the best hit? Compare the alignments of first and second hits. Which one do you think matches the best? Why? 2 2015-12-10 9. Do you find the same structure using BLAST and FASTA? Compare the scores and Evalues obtained from BLAST and FASTA. PSI-BLAST (Position Specific Iterated BLAST) Go to BLAST home page and choose protein BLAST. Use the same protein sequence as a query and choose PSI-BLAST as Algorithm. And BLAST it. 1. When you get results, have a look at the top hits. The result is the same as for the normal BLAST. What is the last hit? Is it significant? 2. Note that all the hits are tick marked on the right. These sequences will be used to build a Position Specific Scoring Matrix (PSSM) (a scoring matrix similar to BLOSSUM and PAM matrices you saw in the slides). Now run next iteration of BLAST by clicking GO below the hit sequences. Did you notice that you can limit the number of sequences in the next iteration? If you did not notice, find the option in the next iteration. 3. When you get the results, scroll down to the hit sequences. Do you find any sequences highlighted on yellow? If there are some, these are the new sequences found in the second round. 4. Now, go the end of the hit sequences and run next iteration. You will find some more new protein sequences in the results. 5. Perform 5 iterations of PSI-BLAST. Do you still see new sequences found in the last iteration? Why do you think there are new sequences in each iteration? Email your reports to: abhishek.niroula@med.lu.se 3