Supplementary Fig. 1. Targeted disruption of Cc.arp9. (A) A diagram
advertisement
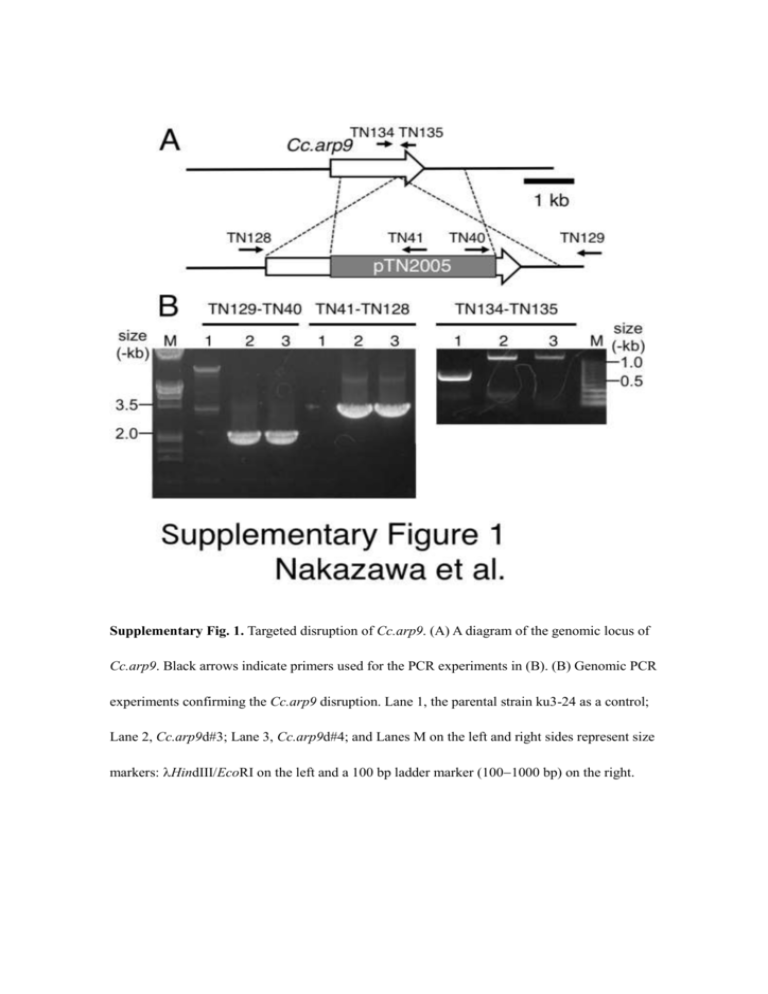
Supplementary Fig. 1. Targeted disruption of Cc.arp9. (A) A diagram of the genomic locus of Cc.arp9. Black arrows indicate primers used for the PCR experiments in (B). (B) Genomic PCR experiments confirming the Cc.arp9 disruption. Lane 1, the parental strain ku3-24 as a control; Lane 2, Cc.arp9d#3; Lane 3, Cc.arp9d#4; and Lanes M on the left and right sides represent size markers: HindIII/EcoRI on the left and a 100 bp ladder marker (1001000 bp) on the right. Supplementary Fig. 2. Isolation of a nucleus in which marker excision occurred through 5-FC counterselection and PCR confirmation. Protoplasts prepared from mycelial cells of strain Cc.arp9d#3 were subjected to 5-FC counterselection to generate strains in which the Cc.arp9 disruption construct was removed. After the counterselection, 11 5-FC-resistant strains were isolated. Nine of the 11 strains exhibited sensitivity to hygromycin B, suggesting that the construct used for Cc.arp9 was removed through intramolecular homologous recombination. Genomic PCR using two, Cc.arp9d_rev#1 and Cc.arp9d_rev#2, of the nine strains, as well as Cc.arp9d#3 and ku3-24 as controls, were then performed. (A) Diagram of the genomic locus of the Cc.arp9 gene in strain ku3-24, Cc.arp9 disruptants from ku3-24 (before 5-FC counterselection) and strains in which the Cc.arp9 disruption construct was removed (after 5-FC counterselection). Thin arrows indicate the primers used for PCR. Thick arrows indicate direct repeat sequences that mediate intramolecular homologous recombination to remove expression cassettes for Hph and PoFcy1. (B) Genomic PCR experiments confirming that marker excision occurred. Lane M, size markers (HindIII on the left side, and HindIII/EcoRI on the right side); Lane 1, Cc.arp9d_rev#1; Lane 2, Cc.arp9d_rev#2; Lane 3, Cc.arp9d#3; and Lane 4, ku3-24. The reason for the amplification of small amounts of three fragments, indicated by white arrows and were expected to be amplified from the genomes of strains in which the construct was removed (after 5-FC counterselection), from genomic DNAs from strain Cc.arp9d#3 (before 5-FC counterselection) is described in Nakazawa and Honda (2015). Supplementary Fig. 3. Generation of Cc.arp9d/Cc.snf5d double-gene disruptants by introducing the Cc.snf5-disrupting construct into Cc.arp9d_rev#2. (A) Diagram of the genomic locus of the Cc.snf5 gene. Arrows indicate the primers used for PCR experiments in (B). (B) Genomic PCR experiments confirming that the Cc.arp9d/Cc.snf5d double-gene disruptants had been generated. We designated this mutation Cc.snf5-2. Lane1, Cc.arp9d/Cc.snf5d#4; Lane 2, Cc.arp9d/Cc.snf5d#5; Lane 3, Cc.arp9d_rev#2; Lane 4, ku3-24; and Lane M, HindIII size marker. Supplementary Fig. 4. The effects of the Cc.arp9-1 mutation on oidiaphore development. Selected micrographs showing oidiaphores (white arrows) from indicated strains, which shows that the number of mature oidiaphores produced in Cc.arp9d#3 is less than that produced in its parental strain ku3-24. For more details, please see Table 4. Scale bars represent 25 µm. Supplementary Table 1. Primers used in this study. Primer Sequence (5’ - 3’) TN40 ACCCTTTCCCCCAAAATTTGGAAGC TN41 ACCTTCTGGCATGACCTTTTGATGATCGC TN95 TACAACGAGATCGGTGCCAGC TN96 AAGCCGGTCATGAAGAAGTGGAGACG TN116 AAATACGTGCTCTTCAACCAACTTCAACG TN117 ACGTCGTGACTGGGAAGAAGACACAACGCCATCAG TN118 CCTGTGTGAAATTGTTCTTCAACGACAACAGCCAG TN119 ACGATGGAACAACAGGTTGGTGG TN128 ACCGTCGTACGTGTATTCCCAGC TN129 TGGTGGCGAGGGTGGTCAACTAG TN134 TCTGACGCGAAGCTCTTCCACC TN135 AACCGCATCCTTGTCATCACGG TN164 TATGGGCATGGGAGGTGGAAAC TN165 ACGTCGTGACTGGGAAGTTCTTCCAGGCCGAGGTG RN166 CCTGTGTGAAATTGTAGGGTATCAAGAGGGAGGGG TN167 TGGTCATCAAGTTTCATGGAACCGAC TN168 TGCGAGATTTCGGTTGGGACTC TN169 TCTGTGAGATGACCAACGAGGAAGAG TN170 TTACGATCCCGAAGCGTGCC TN171 AAGAGGAACGAATGGGCTTTGAAG TN197 AAGCCAGCCATCCCTTCAGC TN198 TGCGAGATTTCGGTTGGGACTC TN250 TCTGGGTTCACAGCCATGAGAGC TN251 ATGCCAGTTCTGAGACAGCACGTTC M13F TCCCAGTCACGACGTTGTAAAACGACGG M13R ACAATTTCACACAGGAAACAGCTATGACC arp9_5RACE-1 ACCAGCTGCTTCGCCAGTCG arp9_5RACE-2 TTGCATCACGCTCGAATTGCT arp9_3RACE-1 TGCTCTTCAACCAACTTCAACGACG arp9_3RACE-2 AACGGTTCAATGTACCCGCATTCG Supplementary Table 2. Effects of the Cc.arp9-1 mutation on clamp cell formation. Strain 28ºC 37ºC ku3-24 99.2% (126/127) 1 100% (100/100) 1 Cc.arp9d#3 100% (105/105) 1 99.4% (174/175) 1 Cc.arp9d#4 99.1% (114/115) 1 100% (155/155) 1 Values in parentheses are “no. of septa with clamp cells / no. of septa observed”. For the effects of Cc.snf5 disruption 1 (Cc.snf5-2) on clamp cell formation, please see Ando et al. (2013).