Abstract Duplications junctons reveal new classes of dispersed
advertisement

ABSTRACT
Duplications junctons reveal new classes of dispersed repeats in the
Salmonella chromosome.
Eric Kofoid, Laszlo Csonka and John Roth
Department of Microbiology and Molecular Genetics, UC Davis, Davis, CA 95616
{ERICs: There are 13 nearly full-length matches. Eliminating those, there are 25
more than half full-length. There are many less than full length which still have a
compelling alignment.}
Eukaryotic chromosomes contain large amounts of DNA in the form of
amplifications of small to medium-sized elements whose number is variable from
organism to organism and even from cell to cell. This often suggests detritus
tolerated by a system with little need for streamlining.
The case for prokaryotes is distinctly different. Their genomes tend to be compact
reservoirs of information. Genetic elements which are repeated and distributed
throughout the genome must be under selection to survive the drive towards
compaction, either because their multiplicity benefits the host as a whole, or
because their selfish nature punishes the host when their number is depleted.
Such elements can be clustered as tandem amplifications or dispersed throughout
the chromosome. These are not mutually exclusive states, as they can also be
dispersed as multiple clusters. Their presence and distribution are often sufficiently
stable to serve as taxonomic markers for classification.
Chromosomal duplication of regions that are not flanked by prominent direct
repeats (e.g rrn sequences) arise at high frequency in a manner that does not
depend on either early recombination functions (RecA, RecBC, RecFOR) or late steps
(RuvC, RecG). Some may arise by palindrome processing with intermediate tandem
inversion duplications. Others may form by single strand annealing between short
sequence repeats. The high frequency of these duplications is likely to reflect the
abundance of short sequence pairs that can serve individually as low-frequency
exchange points (Deletion frequency is restricted by proximity of essential genes.)
Paralogous functional genes provide duplication junctions only rarely. In
Salmonella LT2, many RecA-independent duplications form between dispersed
short sequence repeats that are or may once have been transposable.
Repeated elements containing actual translated genes are generally easy to
rationalize. Some, such as the 7 ribosomal RNA operons of E.coli or Salmonella
simply reflect the need for an abundance of certain products -- in the case just
named, ribosomes -- to fulfill metabolic requirements of the cell. Others, such as the
several IS elements in many bacteria, exploit parasitically the resources of the cell
and find strength in numbers. Our interest is in those entities lacking genes. Are
they parasites? Do they subserve regulatory functions such as DNA-protein
interaction sites? Do they encode ribozymes? Are they used for nucleoid assembly,
gyrase cutting, DNA replicase modulation, or other DNA-level activity?
Several such elements have been found across a number of bacterial clades. Some
which also characterize most Salmonella are the following. REPs (Repeated
Extragenic Palindromic sequences; Stern 1984) are examples of dispersed clusters.
Individually, they are about 40 nt long but are frequently in clusters, some as long as
12 elements (Blattner, 1997). They are known to be mobilized in trans by specific
transposases, suggesting they are domesticated insertion sequences (Ton-Hoang
2011). "Box C" elements (Bergler 1992) are quasipalindromes 56 nt long, occurring
in small arrays. Many copies are found in the E.coli chromosome, but only one
occurs in Salmonella. Transcripts crossing Box C elements can interact with nucleoid
protein HU (Macvanin 2012), but the significance of this is unknown. CRISPR
(clustered regularly interspaced short palindromic repeats) are 25-50 nt clustered
elements which enter the cell by horizontal transmission and interact with Cas
(CRISPR-associated) proteins to provide the cell with an effective adaptive immune
response (Bolotin 2005).
We describe here two new repeated elements in Salmonella enterica typhimurium
and present evidence that further repeated elements in this taxon are unlikely to be
found. One class, "Aelreps", has distinctive features characteristic of MITEs
(Miniature Inverted Repeat Transposable Elements; Correia 1996). Twelve copies
occur throughout the chromosome, 7 of which are full size (222 nt). The other class,
"Lasreps", are found 8 times full-size in the chromosome, and many partial
examples also exist. They do not resemble any other known class of repeat
elements. Both types of elements are widely dispersed in most Salmonella species as
well as in related enteric taxa.
We used an exhaustive computational search tool (the Piler suite; Edgar 2005) to
locate all repeated sequences numbering 3 copies or more of size 50-2000 nt in the
LT2 genome. We feel that the two repeats described in this paper complete the
inventory of such elements in this organism.
============ Notes from John ===========
Chromosomal duplication of regions that are not flanked by prominent direct
repeats (e.g rrn sequences) arise at high frequency in a manner that does not
depend on either early recombination functions (RecA, RecBC, RecFOR) or late steps
(RuvC, RecG). Some may arise by palindrome processing with intermediate tandem
inversion duplications. Others may form by single strand annealing between short
sequence repeats. The high frequency of these duplications is likely to reflect the
abundance of short sequence pairs that can serve individually as low-frequency
exchange points (Deletion frequency is restricted by proximity of essential genes.)
Paralogous functional genes provide duplication junctions only rarely. In
Salmonella LT2, many RecA-independent duplications form between dispersed
short sequence repeats that are or may once have been transposable.
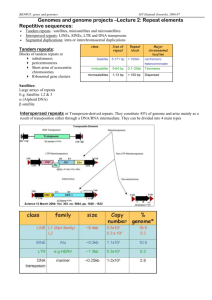
These include REP elements (30bp repeats; 600 copies), IS200 sequences (700bp
repeats; 6 copies) and Eric sequences (100bp imperfect repeats; ### copies).
These three elements seem too short to be good substrates for homologous
recombination. To our surprise, one duplication of the pyrD gene arose by
exchanges between copies of a new element (Aelric) -- 100bp in ## copies. This
reminded us another such element discovered by L. Csonka (Laszlo); 100bp and ##
copies. Apparently both elements had been missed by annotators, and our
searches suggest that these are the last two of comparable size and similarity.
Aelric is found primarily in Salmonella, but occasionally in other enterics
where the copy number is lower and the sequences more widely divergent from the
LT2 consensus sequence. Laszlo is distributed more widely but…….. Both Aelric and
Laszlo like REP and ERIC???) seem to be members of a general class of elements
(MITES) in which the frequently encountered copies do not include a transposase,
but are associated in some genomes with a transposase gene that is capable of
catalyzing their transposition. The transposase of IS200 may be related to the
parent transposase of REP elements. A likely parent transposase for Aelric has
been identified in E.coli, but none has been suggested for Laszlo. The LT2 copies of
both elements are closely conserved (differ by ???); There is evidence that (like
REP) the ends of these elements are more conserved than the centers.