- ClinGen
advertisement

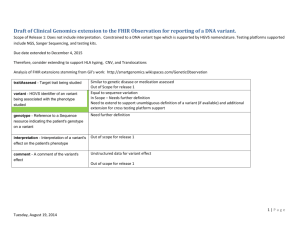
ClinGen Aggregated Sequence Variant and GeneInsight IRB Template Title: Genomic Data Sharing to Improve Health Sponsor Name: NIH Purpose Genomic variation underlies almost all human disease. Technological advances are quickly making variant detection across the genome commonplace in the medical care environment, sparking an expansion of both basic and clinical research. At this time, however, our ability to detect DNA variation has greatly surpassed our ability to interpret the clinical impact of these variants, particularly given the lack of publicly available, carefully curated information on variants and phenotypic consequences. Although collections of some disease-associated variants are available in certain online mutation databases, the variant annotations are limited, often inaccurate and subject to inconsistent standards. In contrast, carefully curated disease-associated variants from patient populations evaluated by individual clinical laboratories are often sequestered in internal laboratory databases making them unavailable to the community. To address these challenges, two efforts are underway: 1) The GeneInsight software will be networked through point to point and share and share alike linkages to enable laboratories to share useful data for the clinical interpretation of genomic variants and 2) the National Center for Biotechnology Information (NCBI) in collaboration with the Clinical Genome Resource (ClinGen) has created the ClinVar database of clinically annotated genomic variation that concentrates the knowledge and curation capabilities of our clinical laboratory community into a public environment. Sharing data from the large number of clinical genetic tests being performed on patients with disease phenotypes presents a unique opportunity to contribute to our understanding of the functional significance of human genomic variation, which will benefit the growing community of medical genomics clinicians and researchers. The investigators at LAB NAME HERE will focus on collecting and sharing information on genomic variants identified through clinical genetic testing in the LAB NAME HERE. The specific objectives of the GeneInsight and ClinGen projects are: 1) Support a standardized infrastructure for data acquisition, storage and clinical use within GeneInsight, a laboratory variant database. 2) Coordinate the submission of aggregate variant data from GeneInsight into ClinVar, a unified database at NCBI. 3) Enable sharing of variant and phenotypic data between laboratories within the GeneInsight network. 4) Implement sustainable expert clinical level curation systems for human genomic variants. Study Population Patients who have had clinical genetic testing through LAB NAME HERE. This includes patients with genetic disorders as well as patients with somatic cancer and patients with a family history or other risk for disease (edit this section as appropriate for your laboratory). Data To Be Collected / Obtained (edit this section as appropriate for your laboratory) Administrative: Coded encounter data (diagnoses, procedures, dates) Demographic data (age, gender, vital status) Personal data (name, address, PCP) Health / Medical: History / Physical Problem List Health/Medical Reports/Results: Laboratory Pathology reports (reports only). Radiology Other Health/Medical Information: Clinical genetic testing results Protected (Identifiable) Health Information PHI refers to health/medical information that is accompanied by any of the listed 18 HIPAA identifiers or by a code where the key to the code that links to the identifiers is accessible to investigators. DE-IDENTIFIED DATA (without any identifiers or codes that link back to individuals) are not considered PHI, and are not subject to HIPAA regulations. Will you be recording any of the identifiers listed above with the data or using a code to link the data to any of the identifiers? If yes, than under the HIPAA Privacy Rule provisons the data cannot be considered de-identified and authorization from the subject or a waiver of authorization must be granted by the IRB. When answering this question, consider the need for recording dates or retaining direct identifiers, such as name and/or medical record number, to link data from multiple sources, to avoid duplicating records, or for QA purposes. NOTE: If you are recording medical record number or other identifiers, even if temporarily for QA purposes or to avoid duplicating records, then answer "Yes". Check the identifiers that will be recorded with or linked by code to the data (edit this section as appropriate for your laboratory). Name Medical record number Dates (except year), e.g., date of birth; admission / discharge date; date of procedure; date of death Other identifier; or combination of identifiers likely to identify the subject will be recorded: Lab accession number and family number Explain why it would be impossible to conduct the research without access to and use of identifiable health / medical information. Because GeneInsight serves as a clinical laboratory system, it must contain PHI. However, no PHI will be visible within the GeneInsight network nor will it be submitted to NCBI. Explain why identifiers must be retained indefinitely, for example, for a health or research purpose, legal or institutional requirement, or other reason. Be specific. PHI must be maintained as the GeneInsight system is also used for clinical care and must therefore be retained according to CLIA guidelines. Waiver of Informed Consent / Authorization The risks for this project involve the release of identifiable clinical or genotype data. Multiple protections are built into the program to reduce this risk. The investigators recognize that rights and privacy of subjects who participate must be protected at all times. Therefore, care will be taken to ensure that data shared within GeneInsight Network and submitted to NCBI will be free of identifiers that would permit linkages to individual subjects and free of data elements that could lead to disclosure of individual subjects. The GeneInsight Suite is registered with the FDA as a Class 1 exempt medical device and as such adheres to a strict quality assurance process. All GeneInsight releases are run through Veracode static analysis and are not released until they achieve a score consistent with Partners security policies. End user activity within GeneInsight is logged and the ability to determine which users have viewed which patients is tested before each major release. Case IDs are created by GeneInsight to allow an outside viewing laboratory to uniquely note and go back to case data examined to support clinical interpretations; however, those IDs will be separate from the labs accession IDs. Language will be added to LAB NAME HERE requisition form to notify patients and providers that deidentified information may be shared with other clinical laboratories. Data from LAB NAME HERE submitted to NCBI's ClinVar will consist of only variant-level data, including variants and their annotations with evidence aggregated across one or more individuals. In order to submit data to ClinVar, data will initially be exported from GeneInsight by GeneInsight staff and later through the development of a computer interface between GeneInsight and ClinVar. GeneInsight creates unique IDs for the variants. Variant IDs are important for enabling data to be updated over time in ClinVar. Variant IDs are separate from the lab accession IDs (which will not be shared with ClinVar). The data elements of ClinVar are stored using reliable, standard technologies, including a relational database in order to ensure transactional integrity, backup, transaction history and rollback, and custodianship. Deidentified, coded data will be securely transferred to the databases at NCBI under appropriate data security protocols. Re-contacting patients to obtain formal consent is not possible for most clinical laboratories and not feasible for the scope of this project. Often, patient contact information is not provided to clinical testing laboratories, making it impossible to contact patients directly. Multiple measures are in place to protect the privacy of individuals' and their health information. Any data shared within the GeneInsight Network will be free of identifiers linking individuals to the data. Data shared with ClinVar will be aggregate variant-level data. The following Privacy Safeguards will be in place for all publicly shared aggregate variant-level data in NCBI's ClinVar: 1) All data samples will include the name of the testing laboratory and a laboratory provided variant code. This code will not be displayed with the data, a separate ClinVar ID will be publically displayed with the variant. 2) No personal health identifiers for the tested individuals or information regarding the ordering physicians will be included in the data submission. 3) NCBI will never be provided with the keys to the codes. Research Data Electronic Research Data (edit this section as appropriate for your laboratory) The desktop computers that the research data will be stored and accessed on are located in the LAB NAME HERE offices. The servers that house GeneInsight are secured within the Partners Needham data center. ClinVar is a separate entity and is maintained by NCBI. Who will have access to the electronic research data? Study staff who are members of LAB NAME HERE will have access to the research data. These study staff members are part of the LAB NAME HERE staff and are involved in the routine operations of clinical genetic testing and support for the interpretation of test results. Their access to the data will follow the protocols of the clinical testing laboratory. Those not working directly with LAB NAME HERE will not have access to identifiable information in the LAB NAME HERE GeneInsight instance, but rather access to the larger data set available through GeneInsight Network (through a participating laboratory) or the aggregate variant-level data available to the public through ClinVar. Sending Health / Medical Information to Collaborators Outside LAB NAME HERE No data with direct identifiers will be shared outside LAB NAME HERE. De-identified caselevel health and medical information related to genetic test results (listed on the "GeneInsight data fields" data collection form, attached) may be accessed by members of the GeneInsight Network, but will not be directly sent to collaborators outside LAB NAME HERE. Aggregate variant-level data (listed on the "GeneInsight data fields" data collection form, attached) will be sent outside LAB NAME HERE to NCBI but will not contain health or medical information on any one individual. Deidentified health information may be accessed through GeneInsight Network by members of participating laboratories. Only aggregate variant-level data will be sent outside LAB NAME HERE, to NCBI for the ClinVar database.