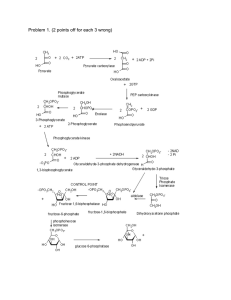
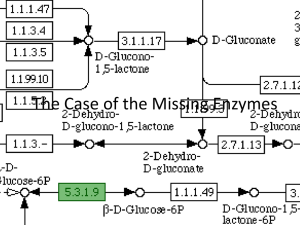
pro2818-sup-0001
advertisement

3 Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenes 4 A versatile dehydrogenase oxidizing alcohols and carbohydrates 1 2 5 6 Henriëtte J. Rozeboom1, Shukun Yu2*, Rene Mikkelsen2; Igor Nikolaev3; Harm J. Mulder3 and 7 Bauke W. Dijkstra*1 8 9 1 Laboratory of Biophysical Chemistry, Groningen Biomolecular Sciences and Biotechnology 10 Institute, University of Groningen, Nijenborgh 7, 9747 AG Groningen, The Netherlands 11 2 12 and 3Archimedesweg 30, 2333 CN Leiden, The Netherlands 13 * 14 e-mail: b.w.dijkstra@rug.nl Phone +31 50 363 4381 / 4378, Fax +31 50 363 4800 or 15 shukun.yu@dupont.com 16 Running title: structure of quinone-dependent alcohol dehydrogenase DuPont Industrial Biosciences, Edwin Rahrs Vej 38, DK 8220 Brabrand, Aarhus, Denmark To whom correspondence may be addressed: 17 18 1 1 Figure S1 Multiple sequence alignment. Structure–based alignment of PQQ-ADH from 2 Pseudogluconobacter saccharoketogenes IFO 14464 (4CVB), L-sorbose dehydrogenase SDH 3 from Ketogulonicigenium vulgare 4 ADHIIG (1YIQ)2 and ADHIIB (1KV9)3 from Pseudomonas putida HK5, QH-ADH from 5 Comamonas testosteroni (1KB0)4, QADH from Pseudomonas aeruginosa (1FLG)5 and MDH 6 from Methylacidiphilum fumariolicum SolV (4MAE)6, Methylococcus capsulatus (4TQO)7 7 Methylobacterium extorquens (1W6S) 8, Hyphomicrobium denitrificans (2D0V)9 Paracoccus 8 denitrificans (1LRW) 9 made with Promals3D 12. The secondary structure elements above the sequence alignment are 10 those obtained from the crystal structure of PQQ-ADH. Residues involved in Ca2+ binding 11 have a purple background color, identical residues have a red background color and similar 12 residues have a red color. Residues indicated with blue stars below the sequences are PQQ 13 ligands and with grey stars above the sequence have alternate conformations. The vicinal 14 disulfide bridge has a cyan background. Green boxes indicate the tryptophan docking motifs. 15 Disulfide linkages are indicated in green italics below the sequences. Residues not in the 16 model of SDH are colored orange. The figure was created with ESPript 13. 17 Figure S2 Overlay of the structures of PQQ-ADH with calcium and zinc bound in the 18 active site. In green the structure is shown with calcium and in cyan with zinc bound. The 19 calcium ion is shown as a green sphere and the zinc ion as a brown sphere. 20 Figure S3 Stereo image overlay of the structures of PQQ-ADH (in green) and QH-ADH 21 (in magenta). The PQQ cofactor is shown in grey sticks and the heme cofactor in yellow 22 sticks. Cytochrome c551 from (PDB ID 351C) 23 extorquens (PDB ID 2CS8) 15 superposed on the cytochrome domain of QH-ADH are shown 24 in cyan and blue respectively. 10 (4MH1)1, quinohemoprotein alcohol dehydrogenases and Methylophilus W3A1 (4AAH)11. The structural alignment was 14 and cytochrome cL from Methylobacterium 25 2 1 References 2 1. Han X, Xiong X, Jiang D, Chen S, Huang E, Zhang W, Liu X (2014) Crystal structure of L-sorbose 3 dehydrogenase, a pyrroloquinoline quinone-dependent enzyme with homodimeric assembly, from 4 Ketogulonicigenium vulgare. Biotechnol Lett 36:1001-1008. 5 2. Toyama H, Chen ZW, Fukumoto M, Adachi O, Matsushita K, Mathews FS (2005) Molecular 6 cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADH-IIG from 7 Pseudomonas putida HK5. J Mol Biol 352:91-104. 8 3. Chen ZW, Matsushita K, Yamashita T, Fujii TA, Toyama H, Adachi O, Bellamy HD, Mathews FS 9 (2002) Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas 10 putida HK5. Structure 10:837-849. 11 4. Oubrie A, Rozeboom HJ, Kalk KH, Huizinga EG, Dijkstra BW (2002) Crystal structure of 12 quinohemoprotein alcohol dehydrogenase from Comamonas testosteroni - Structural basis for 13 substrate oxidation and electron transfer. J Biol Chem 277:3727-3732. 14 5. Keitel T, Diehl A, Knaute T, Stezowski JJ, Hohne W, Gorisch H (2000) X-ray structure of the 15 quinoprotein ethanol dehydrogenase from Pseudomonas aeruginosa: Basis of substrate specificity. J 16 Mol Biol 297:961-974. 17 6. Pol A, Barends TR, Dietl A, Khadem AF, Eygensteyn J, Jetten MS, Op den Camp HJ (2014) Rare 18 earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16:255-264. 19 7. Culpepper MA, Rosenzweig AC (2014) Structure and Protein-Protein Interactions of Methanol 20 Dehydrogenase from Methylococcus capsulatus (Bath). Biochemistry (N Y ) 53:6211-6219. 21 8. Williams PA, Coates L, Mohammed F, Gill R, Erskine PT, Coker A, Wood SP, Anthony C, Cooper 22 JB (2005) The atomic resolution structure of methanol dehydrogenase from Methylobacterium 23 extorquens. Acta Crystallogr D 61:75-79. 24 9. Nojiri M, Hira D, Yamaguchi K, Okajima T, Tanizawa K, Suzuki S (2006) Crystal structures of 25 cytochrome cL and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and 26 mechanistic insights into interactions between the two proteins. Biochemistry 45:3481-3492. 27 10. Xia ZX, He YN, Dai WW, White SA, Boyd GD, Mathews FS (1999) Detailed active site 28 configuration of a new crystal form of methanol dehydrogenase from Methylophilus W3A1 at 1.9 A 3 1 resolution. Biochemistry-Us 38:1214-1220. 2 11. Li J, Gan JH, Mathews FS, Xia ZX (2011) The enzymatic reaction-induced configuration change 3 of the prosthetic group PQQ of methanol dehydrogenase. Biochem Biophys Res Commun 406:621- 4 626. 5 12. Pei J, Tang M, Grishin NV (2008) PROMALS3D web server for accurate multiple protein 6 sequence and structure alignments. Nucleic Acids Res 36:W30-4. 7 13. Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence 8 alignments in PostScript. Bioinformatics 15:305-308. 9 14. Matsuura Y, Takano T, Dickerson R (1982) Structure of Cytochrome-C551 from Pseudomonas- 10 Aeruginosa Refined at 1.6 a Resolution and Comparison of the 2 Redox Forms. J Mol Biol 156:389- 11 409. 12 15. Williams P, Coates L, Mohammed F, Gill R, Erskine P, Bourgeois D, Wood S, Anthony C, Cooper 13 J (2006) The 1.6 angstrom X-ray structure of the unusual c-type cytochrome, cytochrome c(L), from 14 the methylotrophic bacterium Methylobacterium extorquens. J Mol Biol 357:151-162. 15 4