tpj12414-sup-0002-Legends
advertisement

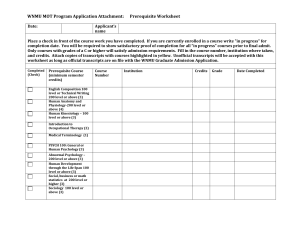
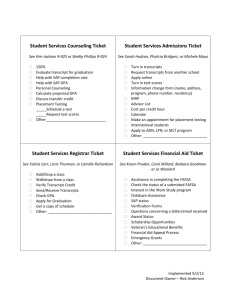
Supporting information Table S1. Highly abundant (expression >median), highly enriched (log fold change >2) celltype specific markers based on the AR to somatic LCM tissue comparison. ZmAGO18a, DYAD/SWI1, a leafbladeless1 homolog and a Cdk2 inhibitor (cell cycle arrest gene), possibly involved in slowing the AR mitotic rate to achieve cell enlargement during early specification (Kelliher and Walbot, 2011), all qualify as highly expressed, highly enriched AR cell markers. An explanation of column labels for this and all subsequent tables follows. ProteinID: GRMZM numbers represents maize gene predictions based on the B73 inbred genome assembly. If GRMZM numbers were not available, the given IDs are from NCBI. Log-fold changes were calculated from the relative cy3:cy5 fluorescence reads across biological and technical replicates, according to the methods described in the manuscript (limma package in R and the lowess method) (Nan et al., 2011). For classification of differential expression, transcripts that were present in one tissue but not another were called as ON/OFF transcripts, which represent a qualitative assessment and thus do not have an associated log-fold change value. In these tables all UP/DOWN transcripts necessarily have a log fold change >0.58 and a p-value <0.05, except in the case of this first table, which has more stringent cut offs (>2). AR Avg. Intensity and Somatic Avg. Intensity: These are averaged intensities across 2 biological and 2 technical replicates after normalization. The Ratio value is obtained by calculating the ratio of the average intensity of one sample to the other. Rank order differences between the Ratio and the log-fold change values reflects the different statistical methods used to obtain them, but in general, confirm the validity of each method and of the classification assigned to each transcript. Table S2. Highly abundant (expression >median), highly enriched (log fold change >2) celltype specific markers based on the somatic to AR LCM tissue comparison. Somatic cells are highly enriched in transcripts for cyclins and numerous kinases putatively involved in cell proliferation and cell-cell communication during the specification of the three somatic cell layers. In addition, other somatic markers include ZmAGO1 family genes such as ZmAGO1b, ZmAGO1d, and ZmAGO10a, all involved in microRNA targeting for RNA interference (by homology to Arabidopsis AGO1 and AGO10 proteins, for which these functions have been ascribed) (Xie et al., 2005). Table S3. Confirmation of cell-type specificity of high quality AR markers with qRT-PCR. These reactions were performed on cDNA made from the same tissue samples as were used for the microarray. qRT-PCR confirmed cell-type specificity for 28/31 germinal transcripts, including two that were classified by qRT-PCR as ON in AR cells and OFF (no amplification) in somatic cells. Seven out of ten AR markers tested by in situ hybridization gave the expected AR-localized pattern, one hybridized to both AR and SPL (see Appendix S1 at the end of this document for more information), and the remaining two (glycosyltransferase and MAM33) did not give any signal. In the “validated?” column, a ‘yes’ indicates confirmation of cell-type specificity by qRT-PCR. The requirement for confirmation was log2 ratio of Ct values had to be >0.58 in the expected direction. This calculation was made from Ct values that were adjusted for primer efficiency using PCR miner (http://www.ewindup.info/miner/version2/) and adjusted for starting cDNA amounts by comparison with the housekeeping gene cyanase. All primers were designed to bridge introns and all passed a gDNA and cDNA test with the expected intron size differences between amplified products analyzed by gel electrophoresis. The three germinal transcripts that failed to confirm array results had log-fold change values <0.58 or were slightly more enriched in the opposite cell type, but the Ct values in each instance were low (>31), at which level the comparisons are inherently less reliable. Also indicated on the table are the array log-fold change values for comparison to qRT-PCR results, and the RNA in situ hybridization result, if that experiment was performed for the given transcript. Table S4. Twenty-four of the 28 somatic transcripts tested by qRT-PCR were confirmed as somatic-specific, including two that were classified in the array as ON in somatic and OFF in AR cells. Eight out of eight somatic markers selected for RNA in situ hybridization to confirm transcript localization gave the expected somatic-specific pattern. See Table S3 legend for more information on the data types in each column. Table S5. AR-enriched or -specific transcripts involved in meiosis. These genes have defined functions in meiosis or were assigned to meiotic progression (Nan et al., 2011) by differential expression in ameiotic1-praI and ameiotic1-489 alleles in meiotic anthers (1.5 mm anther length). The ameiotic1-1 mutant and most other ameiotic1 (am1) alleles in maize have a dramatic phenotype: AR cells appear normal until meiosis, when they conduct mitosis instead. Am1-praI permits meiotic entry but Pollen Mother Cells arrest at the leptotene / zygotene transition, defining the roles of the AMEIOTIC1 protein in two distinct steps of meiosis. Table S6. AR transcripts for genes involved in RNA-binding, ribosome biogenesis, or translation. Table S7. AR-enriched or -specific transcripts for genes involved in alternative metabolic pathways or ROS clean-up. (upper panel) Transcripts encoding proteins that produce cytoplasmic ATP and/or manage reducing equivalents without mitochondrial respiration. In the column on the right side of the table, the metabolic pathway corresponding to each transcript is given. (lower panel) Genes involved in ROS cleanup and managing reducing equivalents, such as superoxide dismutase converting superoxide into oxygen and hydrogen peroxide, antioxidants, glutathione, thioredoxins, peroxidases, and related proteins involved in cell redox homeostasis. Table S8. AR transcripts for genes involved in transcriptional regulation (transcription factors, TFs). The status of these TFs in the msca1 vs. WT array is indicated in the last column as either significantly down-regulated in msca1 compared to WT (“DOWN”) or absent in msca1 and present in WT (“OFF”). Table S9. Somatic transcripts for genes involved in transcriptional regulation (transcription factors, TFs). The status of these TFs in the msca1 vs. WT array is indicated in the last column as either significantly down-regulated in msca1 compared to WT (“DOWN”) or absent in msca1 and present in WT (“OFF”).