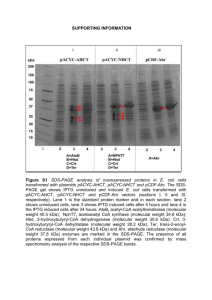
Supplementary methods: ESM_1 Bacterial strains and growth
advertisement

Supplementary methods: ESM_1 Bacterial strains and growth conditions: The bacterial strains and plasmids used in this study are listed in Table 1. Campylobacter strains were routinely cultivated on Columbia agar (GC) supplemented with 5% sheep blood (Oxoid, Dardilly-France) (COS) and the appropriate amount of Campylobacter selective antibiotics supplement (Oxoid) (COS-BW). Growth was generated at 32°C, 37°C, or 42°C as indicated, under microaerophilic conditions (5% O2, 10% CO2, 85% N2) obtained with CampyGen compact (Oxoid). For medium viscosity, bacteria were grown on MH agar or MH liquid medium during 12, 24, 36, 48, 60 and 72h buffered at different pH at 42°C. For osmolarity effect, bacteria were grown either on nutrient broth (NB) (low osmolarity) or on NB supplemented with 20% of sorbitol (NBS) (high osmolarity). Bacterial growth was followed by measuring optical density at 600 nm (OD600). Escherichia coli strains were grown at 37°C on Luria-Bertani (LB) agar or LB medium supplemented with 100 µg/ml ampicillin when indicated. ESM_2 Cloning and expression of Cj1169c: The gene encoding Cj1169c was optimized for E. coli expression and synthesized by GeneCust (Luxembourg). It was subsequently cloned into the pET15b plasmid using NdeI and BamHI restriction enzymes allowing the addition of a sequence encoding 6 histidine residues at the 3’end to help the Cj1169c purification by IMAC (Immobilized-Metal Affinity Chromatography). Two forms of Cj1169c were cloned, one corresponding to the entire coding sequence of Cj1169c and the second one corresponding to Cj1169c deleted of its putative signal sequence, resulting in plasmids pET15b-wtBol and pET15b-mutBol respectively. These two expression plasmids were introduced into E. coli BL21(DE3) by electroporation resulting in SA1 and SA2 strains respectively. Recombinant strains were grown in the presence of 1 mM IPTG at 37°C for 4 h to induce production. Protein production was analyzed by SDS-PAGE and Western blotting using Anti-His antibodies (ESM_5 figure 1). We observed a strong signal at about 8 kDa in the lane corresponding to the truncated Cj1169c and a weaker signal with an apparent molecular weight higher in the lane corresponding to the wild type protein (not shown). While the wild-type form seemed distributed equally in the pellet and in the supernatant after lysis of bacteria, the truncated form appeared mainly in the supernatant (ESM 5, Fig. 1), suggesting that this protein was soluble at least in E. coli. Taking together these observations, we decided to purify the truncated Cj1169c. ESM_3 Production and Purification of Cj1169c: An overnight culture of the SA2 strain was diluted 1:100 in LB and incubated at 37 °C until OD600nm reached 0.6. Protein expression was then induced by the addition of 1 mM isopropyl-β-D-thiogalactoside (IPTG). After a subsequent incubation for 4h at 37°C, the cells were harvested by centrifugation at 10,000 x g for 30 min at 4°C and washed once in Tris-HCl 10 mM, EDTA 1mM pH 7.4. Bacterial cells were disrupted by sonication at 4°C and unbroken cells were removed by centrifugation. Cj1169c purification was performed by IMAC using an Akta Explorer 10, according to the manufacturer’s instructions (GE Healthcare, Velizy-Villacoublay-France). Briefly, the supernatant was loaded on a 5 ml HiTrap-chelating column pre-loaded with NiSO4 and equilibrated with 5 column volume (CV) of the column buffer (Na 2HPO4 20 mM, NaCl 500 mM, pH 7.4) supplemented with imidazole 25 mM, at a flow rate of 2 ml/min. The column was then washed until the optical density at 260 nm reached the baseline. Elution was performed with stepwise gradients using imidazole 75, 250 and 500 mM prepared in the column buffer. The fractions containing pure Cj1169c were collected, pooled and concentrated using an Amicon Ultra-15 Centrifugal Filter Unit (Millipore, Saint-Quentin-en-Yvelines, France) with exclusion limit of 3 kDa, by performing a series of centrifugation at 4,000 x g for 30 min. The concentrated protein was analyzed by SDS–PAGE and used for the production of Cj1169c-specific antibodies. ESM_4 Table 1 Strains and Plasmids used in this study Strains C. jejuni Names Relevant Features Origin Reference NCTC1168 85AF, 85Z, 85S, 87AF, 85X, 85H, 79AH Reference strain Wild-type strains Human Human [6] [1] F38011 LD1 LD2 81176 LD3 Wild-type strain F38011 Δomp50 ΔCj1169c LD1 supplemented with pEKBOmp50 Wild-type strain 81176 Δomp50 ΔCj1169c Human [4] [2] [2] [5] [2] 81176ΔdsbA1 C. coli [3] 85N, 85W, 79K, 87AC, 87AGC LD4 Wild-type strains C. lari 96C15, 96C22 85AB Wild-type strains E. coli BL21(DE3) SA1 SA2 Plasmids pEKBOmp50 pET 15b pET15b-wtBol pET15bmutBol Human 87AGC supplemented with pEKBOmp50 BL21 (DE3) harboring pET15b-wtBol BL21(DE3) harboring pET15b-mutBol Shuttle vector, bearing omp50 operon Overexpression vector, 5.710 kb, Apr pET 15b containing Cj1169c wild-type gene pET 15b containing Cj1169c truncated gene (no signal sequence) [1] [2] Poultry Human [1] Novagen This study This study [2] Novagen This study This study Supplementary figure ESM_5 Figure 1 wt and truncated Cj1169c expression and localization in E. coli strains after bacteria lysis Swt Cwt Smut Cmut Conc wt Cj1169c Truncated Cj1169c Western blotting was developed by anti-His antibodies. Swt, E. coli SA1 supernatant; SMut, E. coli SA2 supernatant; Cwt, E. coli SA1 pellet; CMut, E. coli SA2 pellet; Conc, Concentrated truncated Cj1169c. Supplementary references: 1. Dedieu L, Pagès J-M, Bolla J-M (2004) Use of the omp50 gene for identification of Campylobacter species by PCR. J Clin Microbiol 42:2301–2305. 2. Dedieu L, Pagès J-M, Bolla J-M (2008) The omp50 gene is transcriptionally controlled by a temperaturedependent mechanism conserved among thermophilic Campylobacter species. Res Microbiol 159:270– 278. doi: 10.1016/j.resmic.2008.03.002 3. Grabowska AD, Wandel MP, Łasica AM, Nesteruk M, Roszczenko P, Wyszyńska A, Godlewska R, Jagusztyn-Krynicka EK (2011) Campylobacter jejuni dsb gene expression isregulated by iron in a Furdependent mannerand by a translational coupling mechanism. BMC Microbiol 11:166. doi: 10.1186/14712180-11-166 4. Konkel ME, Garvis SG, Tipton SL, Anderson DE Jr, Cieplak W Jr (1997) Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol Microbiol 24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x 5. Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA (1985) A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J INFECT DIS 152:592–596. 6. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG (2000) The genome sequence of the foodborne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. doi: 10.1038/35001088.