BIO240 Exam #3 SP14 answer key
advertisement
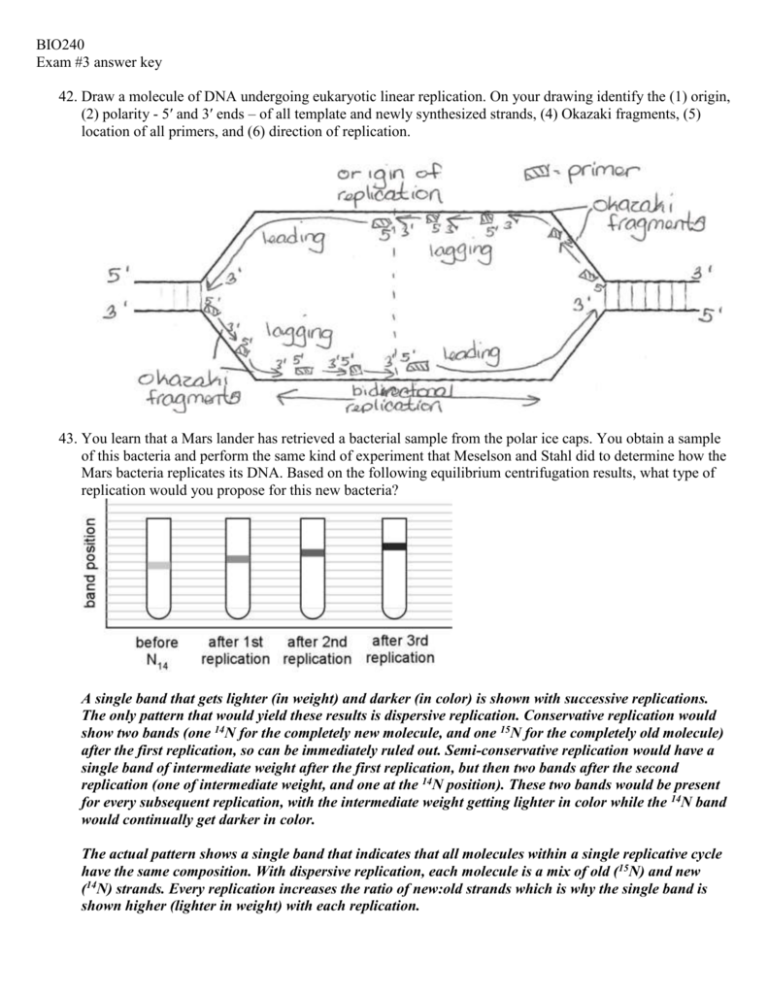
BIO240 Exam #3 answer key 42. Draw a molecule of DNA undergoing eukaryotic linear replication. On your drawing identify the (1) origin, (2) polarity - 5′ and 3′ ends – of all template and newly synthesized strands, (4) Okazaki fragments, (5) location of all primers, and (6) direction of replication. 43. You learn that a Mars lander has retrieved a bacterial sample from the polar ice caps. You obtain a sample of this bacteria and perform the same kind of experiment that Meselson and Stahl did to determine how the Mars bacteria replicates its DNA. Based on the following equilibrium centrifugation results, what type of replication would you propose for this new bacteria? A single band that gets lighter (in weight) and darker (in color) is shown with successive replications. The only pattern that would yield these results is dispersive replication. Conservative replication would show two bands (one 14N for the completely new molecule, and one 15N for the completely old molecule) after the first replication, so can be immediately ruled out. Semi-conservative replication would have a single band of intermediate weight after the first replication, but then two bands after the second replication (one of intermediate weight, and one at the 14N position). These two bands would be present for every subsequent replication, with the intermediate weight getting lighter in color while the 14N band would continually get darker in color. The actual pattern shows a single band that indicates that all molecules within a single replicative cycle have the same composition. With dispersive replication, each molecule is a mix of old (15N) and new (14N) strands. Every replication increases the ratio of new:old strands which is why the single band is shown higher (lighter in weight) with each replication. 44. List and briefly describe the three steps of PCR. Step #1 Denaturation – DNA is heated so that the hydrogen bonds connecting the two strands break. This allows each strand to now serve as a template for a new, second strand. Step #2 Annealing – sample is cooled, allowing primers to form hydrogen bonds to complementary regions of the genome. Step #3 Elongation/Extension – sample is heated. Taq polymerase elongates the new strand started by the primer, adding new nucleotides to the 5’ end of the newly synthesized strand based on complementary base pairing with the template strand. 45. Figure A below shows a restriction map of a rare prokaryotic gene with its direction of transcription indicated by the arrow. Figure B shows the unique cloning region (i.e., multiple cloning site) contained within a plasmid-cloning vector. The blackened region in Figure A represents the amino acid coding sequence of a protein that can be used in humans as a vaccine. The stippled region in Figure B is a highly active, constitutive (unregulated) prokaryotic promoter region. Letters indicate the cleavage sites for different restriction enzymes. Known gene sequences are indicated by short thick lines. Explain how you would isolate and fuse the coding region (Figure A) behind the indicated promoter in the cloning vector (Figure B) to produce large amounts of the protein in bacterial cells. Assume that the cloning vector carries the gene for tetracycline (an antibiotic) resistance. Letters represent different restriction enzymes. Digest your gene region with C and D (you only want to introduce the blackened region, and not other genes). Digest the cloning vector with enzymes C and D, mix with gene fragment C-D, and ligate together using DNA ligase. Transform the ligation products into tetracycline-sensitive bacterial cells and plate out on media containing tetracycline. Incubate the plates overnight. Bacterial colonies growing on these plates the next day should contain the cloning vector and fusion product (i.e., coding regions fused to promoter). 46. Compare and contrast the two basic types of terminators found in bacterial cells in regard to both structure and function. Rho-independent – transcribed RNA contains inverted repeats, which form a hairpin secondary structure. This is followed by a polyA region on the template DNA which will transcribe into a polyU region. The presence of the hairpin slows the speed of RNA polymerase. This reduction in speed, coupled by the weak (2 hydrogen bonds between each A-U base pair) bonds, causes RNA to peel away from its template, and transcription halts. Rho-dependent – transcribed RNA likewise contains inverted repeats, which form a hairpin secondary structure. The following sequence is irrelevant. Rho is an enzyme with helicase activity. Rho protein binds to unstructured (linear) RNA molecule, and travels in 5’ to 3’ direction. The hairpin will slow down RNA polymerase. This allows Rho to “catch up” to where RNA is still hydrogen bonded to its template DNA strand. Rho breaks the hydrogen bonds (regardless of number), causing RNA to be released from its template. 47. The following sequence of nucleotides is found in a single-stranded DNA template: ATTGGCCAGTAATAGGGGCCC Assume the RNA polymerase proceeds along this strand from right to left. a. Which end of the DNA template is the 5′ and which is the 3′? Since polymerase is moving from right to left: 5’ ATTGGCCAGTAATAGGGGCCC 3’ b. Give the sequence and identify the 5′ and 3′ end of the RNA copies from this template. 3’ UAACCGGUCAUUAUCCCCGGG 5’ 48. How do the mRNA of bacterial cells and the pre-mRNAs of eukaryotic cells differ? How do the mature mRNAs of bacterial and eukaryotic cells differ? Bacterial mRNA contains the Shine-Delgarno sequence which serves as a ribosome attachment site. Pre-mRNA of eukaryotic cells contain introns – intervening sequences which are exceedingly rare in prokaryotic cells. mRNA in bacterial cells are mature mRNAs – no further modifications are required, which is why translation of the mRNA can begin before transcription is completed. Mature mRNA in eukaryotic cells have had their introns spliced out, a 5’ guanine cap, and the 3’ end has been cleaved and a polyA tail added.