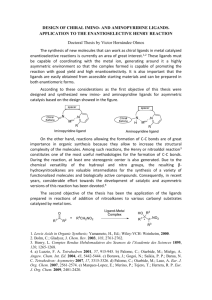
Elucidation of ligand:G-quadruplex structures by NMR - digital
advertisement

OPEN ACCESS DOCUMENT Information of the Journal in which the present paper is published: Bentham Science, Current Pharmaceutical Design, 2012, 18 (14), pp. 1900-1916. DOI: dx.doi.org/10.2174/138161212799958486 1 Experimental Methods for Studying the Interactions between G-Quadruplex Structures and Ligands Joaquim Jaumot* and Raimundo Gargallo Solution Equilibria and Chemometrics group (Associate Unit UB-CSIC), Department of Analytical Chemistry, University of Barcelona, Diagonal 647, E-08028 Barcelona, Spain * Corresponding author Tel: (34)-934034445 fax: (34)-934021233 E-mail address: joaquimjaumot@ub.edu 2 Abstract The present paper reviews the recent advances in and applications of experimental techniques used to study interactions between G-quadruplex structures and ligands that are potentially of pharmaceutical interest. Several instrumental techniques are used to study such interactions. The application of spectroscopic techniques such as molecular absorption, circular dichroism, molecular fluorescence, mass spectrometry and nuclear magnetic resonance are reviewed and we discuss the type of information (qualitative or quantitative) that can be obtained from the use of each technique. Additionally, the application of complementary techniques such as surface plasmon resonance, isothermal titration calorimetry and different methods based on biochemistry is considered. For each technique, the main applications are presented and they are classified according to the family of the ligand and the type of G-quadruplex forming sequence (human telomeric or promoter region of oncogenes) considered. Keywords: G-quadruplex, ligand, review, instrumental techniques 3 Introduction Guanine-rich nucleic acid sequences can fold into four-stranded DNA structures that are known as G-quadruplexes. The building blocks of these structures are G-tetrads or G-quartets: almost planar arrangements of four guanine bases bonded by eight Hoogsteen hydrogen bonds. The G-quadruplex structure can be formed by the intermolecular association of four DNA molecules, by the dimerization of sequences that contain two G-tracts, or by the intramolecular folding of a single strand that contains four blocks of guanines [1, 2]. The existence in vivo of guanine-rich regions at the end of telomeres and near the promoter regions of some oncogenes, such as cmyc, bcl-2 or c-kit is well known [3]. The formation in vitro of G-quadruplex structures in these guanine-rich regions has also been demonstrated. Together, these facts suggest that such structures exist in vivo and that they may be involved in important biological processes, such as aging or the development of cancer [4]. Detailed research has ascertained which factors affect the structure and stability of G-quadruplex structure and the influence of pH, ionic strength, temperature and the presence of organic modifiers are now known. In recent years, particular effort has been made to discover (as well as to design and synthesize) new ligands which that could help to regulate the stability of G-quadruplex structures, thus allowing stabilization or destabilization in vitro [1]. Such ligands should not only modulate the stability of G-quadruplex structures, but they should also do so selectively in the presence of other potential targets, such as double-stranded DNA. Once the ligands are available, a battery of instrumental techniques is used to determine the main characteristics of the G-quadruplex-ligand interactions, such as stoichiometry, mode of binding, and equilibrium or rate constants. The aim of this review is to provide a comprehensive list of the papers published in recent years concerning G-quadruplex-ligand interactions, focusing on the instrumental techniques used in those studies. The published works considered here have been classified according to the instrumental technique employed, and special attention has been paid to those papers that describe novel applications or techniques. Not only have novel techniques been applied to study G-quadruplex-ligand interactions but such interactions may also be the basis for the development of new assays. Previously published reviews Some reviews have been published in recent years that deal with different aspects of G-quadruplex chemistry. Apart from the excellent book edited by Neidle and Balasubramanian [1] that describe the fundamentals aspects of G-quadruplex structures, it is also worth mentioning the volume edited by Baumann [5] in which several methods and protocols are described. The book edited by Fox [6] should also be mentioned: in it several instrumental techniques used to study interactions between nucleic acid structures (not only G-quadruplexes) and drugs are discussed. However, in this section we focus on those reviews that have appeared in journals and that describe the application of instrumental techniques to the study of such interactions. Two different special issues of Methods have been published dealing with the study of the chemistry of quadruplex nucleic acids [7] and, more recently, with the study of nucleic acid structures [8]. In this last issue, several reviews were published of experimental and theoretical methods for studying general DNA or RNA structures, starting from the sequence level, moving through the helical structures and complex tertiary structures, and to their complexes with proteins. However, we are more interested in discuss in depth the special issue devoted to experimental methods used to study G-quadruplex structure. In that issue, the use of probably the most basic experimental method for studying G-quadruplex structures and their interactions with ligands, spectroscopically-monitored melting, was reviewed by Rachwal and Fox [9]. They focus on melting experiments monitored by means of molecular fluorescence, and explained how this can be used to determine the T m values of intramolecular G-quadruplexes and the effects of G-quadruplex-binding ligands. Quantitative analysis of the melting curves can be used to determine the thermodynamic (H, G, and S) and kinetic (k1, k-1) parameters. The method can also be adapted to study the equilibrium between G-quadruplex and Watson-Crick duplex DNA, and to explore the selectivity of ligands for one or other structure. Important problems arising in such experiments and concerning reversibility or choice of fluorophores are also thoroughly discussed. Paramasivan et al. reviewed the applications of circular dichroism (CD) spectroscopy in the study of Gquadruplex structure, as well as the effects on it of cations and ligand binding [10]. The case study approach was used to indicate the strengths and limitations of CD in studies of the properties of G-quadruplex structures formed by telomere repeat sequences, including the determination of the quadruplex structural type, the effect of the sequence on the equilibrium between G-quadruplex structural types, the effect of ligand binding and the effect of 5’-phosphorylation. CD is particularly well suited to monitoring whether changes in cation concentration, the presence of ligands and chemical modification alter the G-quadruplex structure of a 4 DNA in a rapid manner using relatively small amounts of material. Solution NMR spectroscopy has traditionally played a central role in examining G-quadruplex structure, dynamics, and interactions. Webba da Silva provided an overview of the methods currently applied to structural, dynamics, thermodynamics, and kinetics studies of nucleic acid quadruplexes, associated cations and ligand binding [11]. Another technique which has been shown to be successful for studying G-quadruplex-ligands interactions is surface plasmon resonance. Redman reviewed the technique and its main applications in probing G-quadruplex folding and Gquadruplex interactions with proteins and small molecules [12]. In the same issue, the use of competition dialysis in the discovery of selective ligands for G-quadruplex structures was reviewed by Ragazzon and Chaires [13]. Competition dialysis has been shown to be a useful tool for discovering small molecules that selectively recognize the unique structural features of Gquadruplexes as opposed to single-stranded, double-stranded or triplex DNA. The principles and most important practical aspects of this technique were described in detail. Apart from the special issue of Methods, several reviews were published in a special issue of Biochimie concerning the targeting of DNA sequences. The second section of that issue, entitled ‘‘Nucleic Acids Biophysics’’, presented a variety of techniques that may be used to study nucleic acid interactions or properties that may have been overlooked before. The thermodynamic parameters of such reactions may be directly assessed by microcalorimetry (ITC and DSC) which helps to determine the molecular forces involved in the interactions between nucleic acids [14]. A new method has become increasingly popular during the last decade for the study of nucleic acids: mass spectrometry. Gabelica and coworkers presented a review on the state of the art of electrospray mass spectrometry (ESI-MS) in the study of non-covalent nucleic acid complexes [15]. Finally, the fourth section of that issue was dedicated to the ‘‘Structure, biology and targeting of quadruplexes’’. A convenient method for screening Gquadruplex ligands is presented by Teulade-Fichou and colleagues [16]. Several families of compounds are compared using that fluorescence displacement assay, mass spectrometry and FRET melting experiments. Single molecule studies, presented by Shirude and Balasubramanian, may be employed to study the conformational heterogeneity and real time dynamics of these structures [17, 18]. Finally, Neidle and Parkinson [18] reviewed the G-quadruplex X-ray structures published at the time. Most of the crystal G-quadruplex structures described in that work corresponds to sequences from human telomeric DNA or to Oxytricha nova. Since then, several new G-quadruplex crystal structures have been published. Some characteristics of G-quadruplex (loop flexibility and stability of the G-tetrad core) are discussed and related to the few ligand complex structures discussed. So, in this work, our aim is to update the work described above with the new developments and contributions that have appeared in recently. The references included in this work were obtained from a bibliographic search of the Scopus and ISI Web of Science databases in June and July 2011 with a focus on the manuscripts published between 2008 and 2011. However, during the final stages of preparing the manuscript, another review was published covering topics similar to those covered by Murat and coworkers [19] from a different viewpoint that could be considered complementary to our review 5 Experimental Techniques This section contains a list of the different instrumental techniques that have been used to study interactions between Gquadruplexes and ligands. For each technique, we provide a brief theoretical introduction to how it is applied to the study of the interactions, followed by a discussion of some examples that, in our opinion, are characteristic of the technique or that are of specific interest (because they involve a new development or an interesting result). UV-visible Molecular Absorption UV-visible molecular absorption spectroscopy is probably the simplest and most commonly employed instrumental technique for studying both the stability of G-quadruplex structures and their interactions with ligands. Usually, molecules used as ligands show an absorption band that can be clearly distinguished in the visible region. An easy way to determine whether there is any interaction between the G-quadruplex structure and the drug is therefore to analyze the shifting of the position of the maximum of this band from when the ligand is free in solution to when the ligand is complexed with the DNA. It has been postulated that the magnitude of this shifting could be interpreted as an indication of the strength of the interaction between the DNA structure and the ligand considered. In addition to this qualitative information, experiments involving titration of the DNA with the ligand (or vice versa) provide quantitative information such as stoichiometries and binding constants. There are several recent examples in which interactions between the ligand and the G-quadruplex structure are studied by means of a titration monitored by UV-vis molecular absorption spectroscopy [20-55]. Figure 1 near here. Figure 1a shows the shift of the Soret band (at approximately 430 nm) corresponding to the porphyrin TMPyP4 upon successive additions of a G-quadruplex-forming sequence belonging to the k-ras promoter. The dramatic red shift from 422 to 440 nm is an excellent indication of interaction between the G-quadruplex and the ligand. However, obtaining quantitative information is not so straightforward. Stoichiometries and equilibrium constants are calculated by fitting absorbance data to previously proposed interaction models using two different approaches based either on Scatchard plots or on multivariate analysis which provides distribution diagrams and spectra of the species considered (see Figures 1b and 1c). Using these two different approaches the number of species present in the system can be determined and more accurate models for describing interactions between the Gquadruplex structure and ligands may be postulated. This is particularly useful when interactions with different natures are present. Some examples of the application of these data analysis methods are summarized in Table 1. Table 1 near here A different strategy for detecting interactions between G-quadruplex and drugs is to study the thermodynamic parameters calculated from UV-monitored melting experiments. In a melting experiment, the melting temperature and thermodynamic parameters are determined from the changes in the absorbance (usually at 295 nm for G-quadruplex structures) as a function of temperature. Experimental considerations regarding the measuring and analysis of thermal melting curves have been discussed in detail [56]. In this way, comparing the melting temperature values in the absence and presence of a ligand is a tool for easily establishing whether there is an interaction between the ligand and the G-quadruplex. In addition, it indicates whether the complex stabilizes (Tm increases) or destabilizes (Tm decreases) the G-quadruplex structure. An example of the application of melting profiles can be found in the work of Agarwal [57]. In that work, interactions between a sequence based on the c-myc oncogene and two furan-based cyclic homooligopeptides were studied. Figure 2 shows the melting curves recorded at 295 nm of two ligands under four different conditions (in absence of each ligand and at three different ligand concentrations). A clear stabilization effect can be observed for both ligands, but the authors point out that the second ligand increased the melting temperature by more than twenty degrees and showed bigger greater stabilization effect (Figure 2.b). Figure 2 near here. Some examples of other recently published work in which melting profiles are used to study the interactions of G-quadruplexes with several classes of ligand are shown below. They include ligand types such as platinum complexes [32, 43], ruthenium 6 complexes [41, 53], bisbenzimidazoles [37], polyamines [52], oxazoles [24, 58], oxazynes [33], porphyrins and derivatives [30, 31, 35, 38, 48, 59, 60], pthalocyanines [61], perilenes and their derivatives [60], neomycin-perylene conjugates [62] or isoquinoline alkaloids [46]. Circular dichroism Just like UV-visible molecular absorption spectroscopy, circular dichroism (CD) spectroscopy has become an essential technique in the study of G-quadruplex structure and of G-quadruplex-ligand interactions. CD is a straightforward method for monitoring conformational changes in DNA structure induced by modifications of the environment, such as temperature, the nature and/or concentration of counterions, pH or the addition of crowding agents. The effect of chemical modifications on the DNA structure may also be studied by means of CD spectroscopy. It is commonly accepted that the conformation of a G-quadruplex structure (parallel, antiparallel or mixed) can be roughly assigned from the position and magnitude of the CD bands. Hence, a positive band at 260 nm is commonly assigned to a parallel conformation, whereas a positive band at 295 nm and a negative band at 260 are indicative of an antiparallel conformation. However, CD results cannot be used to unambiguously determine the type of quadruplex structure adopted by DNA [10], but it can be used to limit the number of possibilities. In recent work, Masiero and coworkers criticized the paradigm cited above on the basis of a qualitative rationale of exciton coupling between G-tetrads [63]. According to them, the polarity of stacked G-tetrads, and not the relative orientations of the backbone strands, is likely to be the determining factor for G-quadruplex spectra. Hence, the kind of CD spectrum observed would not be directly related to the relative strand orientation: the stacking orientation of the Gtetrads obviously depends on the folding of the strand (and hence on the parallel/antiparallel strand polarity); however, no direct relationship could be established between the two topological features. To sum up, CD should only be used to obtain information about G-quadruplex structures by comparing their spectra with those of known structures by means of NMR or X-ray crystallography. As mentioned above, UV-vis-monitored melting experiments are a common way to obtain information about the stability of a DNA structure. However, as the absorbance hyperchromicity (at 260 nm) or hypochromicity (at 295 nm) of G-quadruplex DNA tends to be much smaller than that associated with duplex DNA, CD has been widely used to monitor thermal unfolding of Gquadruplex structures. This is usually performed by plotting the measured ellipticity at the wavelength corresponding to the maximum of a positive band (around 260 nm for parallel structures or 288 nm for antiparallel structures) versus temperature [6468]). CD spectroscopy has been used extensively to study interactions between G-quadruplex and ligands. In general, these are nonchiral molecules which do not exhibit a CD signal in solution. Hence, changes observed in the UV region when a G-quadruplex structure is titrated with a non-chiral molecule can be directly related to modifications of the G-quadruplex structure. However, in some cases, when a non-chiral ligand binds tightly to chiral DNA, such as a G-quadruplex, a CD signal is induced in the wavelength region corresponding to the absorbance of the bound compound. Intercalating compounds usually produce negative induced CD signals or very small positive signals. Groove binding is generally indicated by the presence of a large positive induced CD signal upon titration of the compound into DNA [69]. In any case and due to the relatively low signal-to-noise ratio, CD is frequently limited to providing qualitative data. In other words, the effect of the ligand on the G-quadruplex structure is merely observed: quantitative determination of stoichiometry and calculation of binding constants require other procedures and techniques, such as UV-vis molecular absorption or fluorescence-monitored titration of a ligand solution with DNA. The human telomeric structure has been the main focus of attention for researches working on the effect of ligands on Gquadruplexes. Therefore, CD-monitored titrations of human telomeric G-quadruplex DNA have been reported with oxazole-based peptide macrocyles [70], perylene-neomycin conjugates [62], disubstituted 2-phenyl-benzopyranopyrimidine derivatives [71], naphathalene diimide derivatives [64], natural alkaloids [72] or their derivatives [46], phenantroline derivatives [73], telomestatin [66], symmetrical bisbenzimidazoles [37], natural polyamines [52], porphyrins [38, 49, 74], metal complexes [41, 48, 53, 75] and a chiral cyclic helicene [76]. Other sequences whose conformational changes upon addition of ligands have been studied include sequences near the promoter regions of genes such as c-kit, c-myc, bcl-2 or vegf. CD spectroscopy was used to study the effects of triaryl- and diethynylpyridine derivatives on a G-rich sequence corresponding to a promoter region of the c-kit gene in the presence and absence of a stabilizing salt [50, 77]. Diethynyl-pyridine derivatives appear to induce the formation of a single parallel conformation of the Gquadruplex. Moreover, the appearance of a relatively weak signal was observed around 390 nm for the ligands, which was explained as a change in the chirality of the proximal chemical environment of the ligand. This sequence has been targeted with 7 oxazole-based macrocycles [70], heteroaromatic 1,4-triazole and with a polycyclic fluorinated acridinium cation [67]. The guanine-rich sequences near the promoter regions of the c-myc gene have also been studied by means of CD spectroscopy. Hence, the effect of polyamines has been studied [52]. Interactions with furan-based cyclic homooligopeptides was qualitatively studied using CD, resulting in the observation that these ligands induced a parallel-stranded conformation [57]. The effect of a berberine derivative has been studied leading to the observation of down-regulation of c-myc gene transcription in the leukemia cell line HL60 [78]. Another G-quadruplex structure studied with CD spectroscopy is that formed by a guanine region near the promoter regions of bcl-2 [39, 52]. To overcome the relatively low signal-to-noise ratio, del Toro et al. used multivariate analysis to analyze spectroscopic data recorded by CD and UV-vis simultaneously monitored titrations of a G-quadruplex-forming region of bcl-2 with TMPyP4 [30]. Finally, interactions between porphyrins and guanine-regions near the promoter regions of vegf [79] and k-ras [80] have also been studied. The interaction of TMPyP4 with k-ras G-quadruplex induces a CD band at 445 nm, which has been explained as a result of end-stacking interaction. Another structure which has been the focus of attention is the hTERT core promoter [68], whose interaction with perylene derivatives has been monitored with CD [65]. Molecular fluorescence Apart from molecular absorption and CD spectroscopy, molecular fluorescence is probably one of the techniques most commonly used to study interactions between ligands and G-quadruplex structures. The advantages of molecular fluorescence over other techniques are its high sensitivity, large linear concentration range and selectivity. The most intense and the most useful fluorescence is found in compounds containing aromatic functional groups with low-energy –> * transition levels. Compounds containing aliphatic and alicyclic carbonyl structures or highly conjugated double-bond structures may also exhibit fluorescence, but the number of these transitions is small compared with those in aromatic systems. When studying G-quadruplex-ligand interactions, there are several different approaches to molecular fluorescence spectroscopy. One consists of including a chemically modified fluorescent base in the DNA sequence of interest, such as, 2-aminopurine which is an analog of adenine that shows fluorescent properties. Nagesh et al. included this base in a DNA sequence corresponding to a bcl-2 promoter region and monitored the increased fluorescence of this base on addition of TMPyP4 [39]. As 2-aminopurine was incorporated into the first and second lateral loops of the G-quadruplex structure, changes in fluorescence seem to support intercalation as the mechanism of the weaker binding mode. A similar approach has been used to explore the binding of calothrixin A to a G-quadruplex from the c-myc gene [81] or oxazole-based macrocyclic compounds to human telomeric DNA [24, 58]. Compounds that have been shown to bind to G-quadruplex DNA have generally been planar, aromatic compounds that bind via external end- stacking to the surface of the G-quartet on either one end or both ends of the quadruplex [69]. As it is found empirically that fluorescence is particularly favored in molecules that possess rigid structures, the fluorescent properties of these ligands may be useful to monitor changes that accompany interaction with G-quadruplex structures. Much research has been performed into the design, synthesis and evaluation of aromatic ligands which can stack on G-quartets. As a consequence, molecular fluorescence spectroscopy has emerged as a useful tool for directly monitoring their interactions with selected DNA sequences. Another way to study the interactions between G-quadruplexes and ligands by means of fluorescence spectroscopy is to use this technique to determine the stoichiometry and equilibrium constants of the complexes formed. In general, experiments involve the addition of aliquots of a stock solution of G-quadruplex to solutions of a ligand. After an equilibration time, the fluorescence spectrum is recorded and the data processed. Once again, the sequence corresponding to human telomeric DNA has been the main focus of attention. Hence, the interaction with a perylene-neomycin conjugate [62], quindoline derivatives [55], isoquinoline alkaloids [46], ruthenium complexes [53], platinum(II) complexes with dipyridophenazine ligands [32] or chiral cyclic helicenes [76] has been described. The interaction of porphyrin-based molecules, such as TMPyP4, which show the rigid structure mentioned above, has been studied for DNA sequences such as human telomeric [34, 38, 45, 49]. Other G-quadruplex-forming sequences which have been studied are those corresponding to several promoter regions. Hence, the interaction of platinum(II) complexes with tridentate ligands with a DNA sequence near the promoter region of c-myc [43] and the interaction of amido phthalocyanines with DNA sequences near the promoter regions of c-myc or c-kit [82] have been described. Moreover, the last is one of the few studies into the pH-dependence of G-quadruplex binding by a ligand. In addition to simple experiments where a ligand is titrated with DNA, comparative fluorescence quenching or polarization anisotropy of the emission spectra may provide additional information about the interaction of G-quadruplexes with ligands, as in 8 the cases of the interaction between amido phthalocyanines and DNA sequences of c-myc or c-kit promoters [82] or the interaction of coralyne and sanguinarine compared to berberine and palmatine [46]. In addition to steady-state measurements carried out to determine equilibrium parameters, kinetic measurements can efficiently be performed with fluorescence instrumentation. In this way, folding induced by potassium ions of oligonucleotides labeled with pyrene at the 5’ and/or 3’ end through a 3-aminopropyl linker has been monitored and the fluorescence decay has been used to determine average lifetimes [83]. These results indicate that even single-labeled pyrene probes could be an alternative in bioanalytical applications. Fluorescence decays can be fitted to exponential curves, thus allowing different binding sites for the ligand to be proposed on the G-quadruplex structure, since longer lifetimes are related to externally bound ligands, whereas shorter lifetimes are related to intercalation or end-stacking [40]. Time-resolved fluorescence spectroscopy has also been used to study the interaction of G-quadruplexes with quindoline derivatives [55] or TMPyP4 [40, 84]. G4-FID (G-quadruplex fluorescent intercalator displacement) is a simple and fast method that allows the affinity of a compound for G-quadruplex DNA and its selectivity towards duplex DNA to be evaluated. This assay is based on the loss of fluorescence of thiazole orange (TO) upon competitive displacement from DNA by a putative ligand under steady-state conditions. The original protocol was tested using various quadruplex (human telomere, TBA) and duplex-DNA targets, and with a wide range of Gquadruplex ligands belonging to different families that it was considered were likely to display various binding modes [16]. However, the original test was limited to the screening of compounds with optical characteristics compatible with those of the probe, i.e., compounds that did not significantly absorb or fluoresce in the same wavelength range as TO. To overcome these difficulties, improvements to the protocol via the use of other fluorescent probes (such as Hoechst 33258 or TO-PRO-3) or by adapting to high-throughput screening have been described [85, 86]. The method has been used to describe qualitatively the interaction of methylene blue derivatives with a G-quadruplex-forming sequence of the c-myc gene [87] and of the aminoglycoside perylene conjugate with human telomeric DNA [62]. Another technique for studying ligand-G-quadruplex interactions which has become very popular is fluorescence (or Förster) resonance energy transfer (FRET) spectroscopy. The DNA sequence to be studied is labeled at the 5’ and 3’ ends with a donor and an acceptor fluorophore. The donor may be 6-carboxyfluorescein (FAM) and the acceptor 6-carboxy tetramethylrhodamine (TAMRA). An excited donor can transfer its energy to the acceptor via an induced-dipole interaction, whereby the efficiency of energy transfer, E, is inversely proportional to the distance between the donor and acceptor. Changes in the distance between the two labeled sites of the molecule can result in a measurable change in the efficiency of the energy transfer. A few years ago, De Cian et al. [88] analyzed the FRET method as used to measure the stabilization and selectivity of quadruplex ligands towards human telomeric G-quadruplex. This was aimed at overcoming some of the drawbacks related to molecular absorbance or CD approaches, such as relatively high DNA and ligand concentrations and related precipitation/aggregation problems, or the absorption of ligands in the region where DNA absorbs. Since the original reports demonstrating that FRET could be used to study G-quadruplexes, FRET systems have become very popular, due to large differences between the fluorescence properties of folded and unfolded forms. The shift in Tm is a measure of the relative strength of the interaction. However, the stabilization value induced by the ligand on the G-quadruplex structure depends on the nature of the fluorescent tags, the incubation buffer, and the method chosen for Tm calculation, which complicates a direct comparison of results obtained in different laboratories [88]. The latest application of FRET melting studies include the study of the interaction of human telomeric DNA with 2-phenylbenzopyranopyrimidine derivatives [71], polyamines [74], substituted naphathalene diimides conjugated to engineered phenol moieties by alkyl-amido spacers [64], triangular aromatic platforms, either with three aminoalkyl side chains (triazatrinaphthylene series), or without side chains (triazoniatrinaphthylene) [89], bis-guanylhydrazone diimidazo pyrimidine [90], substituted acridines [91], and substituted phenanthroline-based compounds [73, 92]. The effect of crowding conditions on ligand-Gquadruplex interactions has also been considered [93]. The interaction of G-quadruplex-forming sequences near the promoter regions of c-myc with berberine derivatives [78, 94], bisaryldiketene derivatives [95], and furan-based cyclic homooligopeptides [57] has been reported. DOTASQ as a free base is relatively ineffective in stabilizing quadruplex-DNA; however, it has been demonstrated that a terbium-mediated conformational switch controls G-quadruplex binding affinity. Thus, the corresponding terbium complexes represent a novel class of quadruplex interacting metallo-organic complexes, interest in which continues to increase exponentially [96]. FRET melting studies have been applied to assess the interaction of quindoline derivatives [97] and polyamines [52] with G-quadruplex-forming sequences near a bcl-2 promoter. Finally, the G-quadruplex formed at the c-kit gene have also been a focus of interest [50, 90, 96, 98]. Without involving a melting process, Green et al. used an original system to study quadruplex opening in the presence of the complementary strand and of a hemicyanine tetrapeptide conjugate by means of FRET [99]. Derived from the classic FRET technique, single-molecule FRET can be employed to directly analyze the conformation of Gquadruplex-forming sequences. Single-molecule detection is a powerful approach for directly analyzing biomolecular properties without the temporal and population averaging that are involved in conventional ensemble studies [17]. As in classic FRET, the 9 biomolecular under study must be labeled with a donor and an acceptor fluorophore. Changes in the distance between the two labeled sites of the molecule can result in a measurable change in the efficiency of energy transfer. Single-molecule FRET has allowed resolution of the intrinsic structural dynamics between the various conformations adopted by human telomeric DNA [100]. Nuclear Magnetic Resonance Nuclear magnetic resonance is also a powerful tool for the analysis of interactions between ligands and G-quadruplex structures. There are two different approaches to its use. Similarly to molecular spectroscopy, NMR can be used to monitor the interaction of oligonucleotides with ligands. Alternatively, NMR can be used to elucidate the structure of the DNA-ligand complex by applying appropriate restrictions to the experimental spectra. In the case of G-quadruplex structures, as described in the 2007 review by Webba da Silva [11], characteristic NMR signals are found in regions of amino and imino protons (between 8 and 13 ppm, approximately) when considering a typical proton spectrum. The NMR spectrum of an intermolecular G-quadruplex structure may show better resolved signals than the spectrum of an intramolecular structure. In the first case, the NMR spectrum of the sequence d(TAGGGTTA) [101] shows only three signals in the imino region corresponding to G3, G4 and G5. In the second case, up to twelve signals corresponding to the twelve guanine bases are observed in the NMR spectrum of the intramolecular G-quadruplex formed by the d(AG3(T2AG3)3) sequence [102]. Higher dimension NMR techniques allow more than one species in equilibrium to be detected and determine their relative populations [103, 104]. Ligand:G-quadruplex titrations by NMR In these experiments the ratio of the concentrations of G-quadruplex and the ligand varies and can be monitored using NMR spectroscopy in the same way as in the case of the molecular spectroscopies discussed above. However, in the case of molecular absorption and CD spectroscopies, the experiments may be performed on-line, i.e., the set of spectra is measured after successive additions of the ligand (or DNA) to the measurement cell. In the case of NMR, the experimental procedure is different and a large number of measurement samples must be prepared. Each sample contains the oligonucleotide under study at a specific concentration (typical concentrations for NMR experiments vary between 0.1 and 10 mM) and a variable amount of ligand to end up with the desired concentration ratios. Interaction between the G-quadruplex and the ligand is confirmed from the comparison of the experimental NMR spectra from all the measurement samples. Typically, the addition of a ligand causes an upfield shift in the characteristic NMR peaks of Gquadruplexes (see Figure 3). This is usually attributed to the disappearance of the signals from the imino protons of the guanines in the G-quadruplex structures and the appearance, upon titration, of signals related to the ligand:DNA bonded species. Moreover, depending on the changes observed in the NMR signals, the binding mode and the strength of the binding between the ligand and G-quadruplex can be proposed. Thus, when the signals corresponding to free and bound DNA are clearly distinguishable, it can be interpreted as tight and specific binding (due to a slow exchange on the NMR scale between free and bound states). However, peak broadening means that a weak and non-specific interaction is present. Finally, depending on whether the shifting of the imino peaks affects all the guanines involved in the tetrads or only those at the ends or central positions, end-stacking or interaction binding modes can be proposed. Figure 3 near here In a recent paper by Wang and collaborators [43], the interaction between an intramolecular G-quadruplex based on the human telomeric sequence (5’ – TGAGGGTGGIGAGGGTGGGGAAGG – 3’) and complexes of platinum and bzimpy derivatives was studied. Upon addition of the platinum complex, some of the NMR peaks (G5, G14 and G20) experienced large shifts and broadening that were explained by the authors as being due to binding of the ligand near the 3’-terminal face of the structure. Pothukuchy and collaborators studied the interaction of TOTA (trioxatriangulenium ion) with several duplex- forming and Gquadruplex-forming sequences [101]. A dramatic upfield shift was observed for the imino NMR signals when studying the sequence d(TAGGGTTA) (Figure 3). As the largest shift was observed for signals corresponding to G5, which is the closest to the 3’-end, interaction of TOTA at this end was proposed. In addition, as the couple of signals corresponding to the free and bound structures were not observed, the authors proposed that there is weak binding between TOTA and the G-quadruplex. The last example considered corresponds to the work of Li et al. [72] in which two non-planar alkaloids (peimine and peimidine) interacted with several G-quadruplex-forming-sequences. The interaction between peimidine and the d(TTAGGGT) 4 oligonucleotide was of particular interest. Upon titration with the ligand, a downfield shift was observed (contrary to the previous 10 examples) of the G4 and G5 imino signals, while the G6 signal did not show any variation. The authors explained this different behavior as being due to a different binding mode between the alkaloid and the G-quadruplex that was finally assigned to a groove binding mode (probably at the side groove in the T2A3G4G5 area). There are several other examples of these types of experiments being applied to the study of G-quadruplex-ligand interactions. For instance, the human telomeric sequence has been studied with perylenes ([105-107], the acridinium salt RHPS4 [108], methylene blue [20], quercetin [21], distamycin A [109], platinum complexes [32], carbazole derivatives [102], amido phthalocyanines [82], acridine and quindoline oligomers [110], natural polyamides [52] and ligands from plant extracts [111]. There are also examples with DNA sequences based on promoter regions of oncogenes such as c-kit, c-myc or bcl-2. The ligands used to study these interactions are diarylethynyl derivatives [112], amido phthalocyanines [82], triarylpyridines [77], acridine and quindoline oligomers [110] and natural polyamides [52]. Elucidation of ligand:G-quadruplex structures by NMR NMR is a powerful tool for elucidating the 3D structure of a G-quadruplex and ligand complex, with its main advantage over crystallography being the fact that the structure is studied in solution and not in a crystal lattice [113]. However, the structure observed is not static but a dynamic image considering all the possible conformations of the molecule. In order to obtain these images, the combination of distance restraints obtained from 2D NMR experiments and other restraints such as torsional angles, trans-hydrogen bonds and dipolar couplings are used. Several examples of G-quadruplex-ligand complexes may be found in the literature. For instance, Martino and coworkers [114] showed the 3D structure of the distamycin A:d(TGGGGT)4 complex combining the data from NOESY experiments and molecular calculations. The structure clearly showed the 4:1 distamycin A:G-quadruplex stoichiometry with two antiparallel distamycin dimers that simultaneously bound two opposite groves of the G-quadruplex structure. Hounsou et al. [115] showed the structure of the complex formed between quinacridines and the d([T 2AG3T]4) sequence. From an NMR titration experiment a 2:1 stoichiometry (quinacridine:G-quadruplex) was derived and a model was proposed with one ligand stacked between A3 and G4 and the other between G6 and T7. Recently, Trotta and collaborators [116] combined NMR and molecular modeling techniques to study the interactions between DNA sequences based on the [d(TGGGGT)]4 oligonucleotide and six molecules obtained in virtual screening that it were expected to interact with the groove of the G-quadruplex structure. In many cases, the authors combined the use of the two different NMR approaches. First, there is an NMR titration in order to discover and confirm the interaction of the G-quadruplex and the ligand and, secondly, the structure of the complex is calculated. For instance, this is the case of the work by Fedoroff [105], Kern [107] and Gavathiotis [108]. Mass Spectrometry In recent years, mass spectrometry (MS) has become one of the most common techniques adopted to study interactions between nucleic acids and ligands. Electrospray ionization (ESI) is the most common ionization method used in the study of biomolecules due to its soft ionization [15, 117-119] in spite of other options such as MALDI ionization [120]. Using ESI techniques, biomolecules can be transferred from the solution to the mass spectrometer with minimal fragmentation and, so, both the mass of the G-quadruplex and the mass of the G-quadruplex-ligand complex can be determined as the non-covalent interactions that formed the complex are not altered during the electrospray process [15]. Focusing on the use of ESI-MS to study complexes, MS gives a signal for each species with a different mass and so it is very straightforward to establish the stoichiometry of the complexes. ESI-MS signals enable several calculations to be performed: the number of DNA strands involved, the number of bound cations (if present) and the number of bound ligands, among others. Taking into account the structure of the nucleic acids, ESI-MS studies are performed using negative polarity. It is well known that the phosphodiester backbone of DNA has a pKa value below 1 and it is fully deprotonated under usual working conditions. In general, in order to preserve their structure, nucleic acid solutions are prepared with monovalent ions. As ESI presents a low salt tolerance, the use of ammonium acetate is preferred because of its good compatibility with ESI-MS. However, it should be taken into account that the topology of the G-quadruplex structure in potassium solution may differ from that adopted in ammonium solution. Moreover, the addition of methanol prior to the injection step may also cause changes in the G-quadruplex structure despite helping to improve the ionization process. Additionally, there are other instrumental settings which must be considered such as capillary temperature, the acceleration voltages in the transfer optics or the lens voltages. In order to have soft conditions 11 in the source (and minimize the possible fragmentation of the complexes), it is recommended that these parameters be kept at the lowest possible values. As stated in the introduction, important work in the application of ESI-MS to the study of G-quadruplex folding and interactions is currently performed by the MS unit at the University of Liege [15, 121-123]. An example of this work is the comparison of the results obtained by ESI-MS with G4-FID and FRET melting for two G-quadruplex-forming sequences (thrombin binding aptamer and human telomeric repeat) and ligands from different families (quinacridines, metallic complexes, etc) [16]. A good agreement was observed for the stoichiometries of the formed complexes obtained by different methods. Casagrande and coworkers reported an extensive study of non-covalent interactions between G-quadruplex structures and several perylene and coronene ligands (telomerase inhibitors) using ESI-MS [124]. They showed that perylene and coronene derivatives formed stable complexes with G-quadruplex oligonucleotides with stoichiometries of 1:1 and 2:1 drug:G-quadruplex complex, as well as calculating the binding constants. Figure 4 near here Li et al. used ESI-MS to characterize the formation of duplex and G-quadruplex structures near the promoter region of the bcl-2 oncogene [60, 125]. In addition, they studied interaction with different ligands and how the structures were stabilized by them. Finally, they concluded that model porphyrin TMPyP4 stabilized the duplex structure (it converts G-quadruplex into duplex DNA, which results in strong and selective binding to the duplex form) while the ligand dehydrocorydaline clearly stabilized the Gquadruplex structure. The stabilization effect of the dehydrocorydaline can be observed in Figure 4. When considering the mixture of the G-rich and C-rich strands, at low concentration of dehydrocorydaline the major species was the duplex. Upon increasing the ligand concentration, the MS signals of the 1:1 and 2:1 complexes between the dehydrocorydaline and the DNA sequence were detected at the same time as the duplex structure was present at a low concentration. Other examples of the application of MS can be found in the literature. The following have been studied by ESI-MS: the interactions of G-quadruplex forming sequences with porphyrins [60, 122, 126, 127], alkaloid derivatives [87, 128, 129], perylene derivatives [60, 122, 125, 127, 130, 131], modified oxazoles [132], triazoles [67, 133], quinacridines [122], polyamides [131], distamycin A [127, 134], daunomycin [129], telomestatin [66, 135], methylene blue [87] and bis-phenantroline derivatives [136]. Surface Plasmon Resonance Surface plasmon resonance (SPR) is a technique that is used to study interactions between G-quadruplex and ligands on the surface of a metal film (usually gold). This technique provides quantitative information about the affinity between the ligand and the DNA and it is thereby possible to determine the stoichiometry, strength and kinetics of the interaction [137, 138]. The current instrumentation allows the interaction of a certain ligand to be studied with several oligonucleotides simultaneously. This makes it possible to compare the binding of a number of different ligands with different quadruplexes quickly and easily [12]. In order to immobilize the G-quadruplex-forming oligonucleotides on the surface of the sensor chips, the DNA sequences are labeled with biotin at either the 5’ or 3’ terminus. The biotin will be linked with the streptavidin that is usually covalently attached to the sensor surface and this will create the biospecific surface. It is important that the folding of the G-quadruplex is maintained throughout the experiment and for this reason experimental aspects, such as the buffer composition, must be taken into account. Finally, the analysis of the experimental sensorgrams requires special care to avoid the effect of non-specific binding or adsorption of ligands to the sensor surface. Recently, Dash et al. [50, 112] showed the usefulness of this technique in the study of the binding of different G-quadruplexforming sequences derived from sequences found near the promoter regions of several proto-oncogenes (k-ras, c-myc, c-kit) with diethynil-pyridine amides. The calculated equilibrium binding constants (K d) for a series of ligands and different sequences were compared. It was shown that appropriate modifications of the parent structure with triazoles and triamides significantly increased the selectivity of G-quadruplex binding. Murat and coworkers [139] compared the effect of G-quadruplex topologies (intermolecular or intramolecular) on the binding affinities with a series of model ligands (TMPyP4, MMQ1, distamycin A, DODC, etc). The sensorgrams and the adsorption isotherms obtained were fitted to a Langmuir model of the TMPyP4:G-quadruplex complex (Figure 5). Using their results, it was proposed that intermolecular G-quadruplex displayed eight-fold selectivity for TMPyP4 compared with the intramolecular structure but, at the same time, the quantity of TMPyP4 bound to the intramolecular structure was larger than that bound to the 12 intermolecular structure. This was also confirmed by the Scatchard analysis of the data; interaction with the intermolecular structure followed a linear model indicating a 1:1 stoichiometry while the interaction with the intramolecular structure followed a nonlinear behavior that was related to the existence of a mixture of parallel/antiparallel structures (or a hybrid 3+1 structure). Similarly, Romera et al. used SPR to determine the selectivity and binding constants for the interaction of several metalloporphyrins with intermolecular, intramolecular (human telomeric) and duplex structures [74]. Figure 5 near here Wu et al. used SPR to determine the equilibrium constants for the binding of disubstituted 2-phenyl-benzopyranopyrimidine derivatives to human telomeric DNA and they compared the results with those previously obtained from FRET measurements [71]. The incubation of these compounds with duplex DNA immobilized on the chips showed either no significant or non-specific binding interactions, which could indicate that the disubstituted derivatives had selectivity for G-quadruplex DNA. Other ligands whose binding to human telomeric G-quadruplex has been studied with SPR include naphthalene diimide derivatives [64], triarylpyridines [140], substituted diarylureas [141], bis(benzimidazole)pyridine derivatives [142], copper corroles [27], zinc phthalocyanine [61], manganese(III) porphyrin [143] and quinolinecarboxamide macrocycle [100]. In addition to equilibrium data, which can be efficiently determined from spectroscopically-monitored titrations, association and dissociation rate constants can be calculated from SPR data. Nielsen et al. determined these values for a series of 4,7-diamino1,10-phenanthroline derivatives binding to human telomeric DNA [144]. In the case of G-quadruplex-forming sequences based on the oncogene promoters, the targeting of c-myc with TMPyP4 [26, 145], furan-based cyclic homooligopeptides [57] or platinum complexes [43] should be noted; as should that of c-kit, also, with TMPyP4 [26]. Finally, in Pillet et al. [146] they proposed the use of a new development of the SPR techniques: SPR Imaging (SPRi). They present SPRi as a technique with the great advantage that multiple determinations can be performed in a single run (up to thousands of spots that can be tested under simultaneous and identical experimental conditions). The results showed that the values of the binding constants determined using this method were consistent with those found in the literature for the sequences (-TTAGGG- human telomeric based) and ligands (a manganese porphyrin, a terpyridine platinum complex, a phenanthrolinebased compound (PhenDC3), and a 2,6-pyridine dicarboxamide derivative) studied. Isothermal Titration Calorimetry Isothermal titration calorimetry (ITC) can be used to measure thermodynamic parameters directly; including the binding affinity constant, enthalpy changes (H) and binding stoichiometry between the ligands and the G-quadruplex structure in solution [147]. From measurements of small changes of temperature, Gibbs energy changes (G) and entropy changes (S) can be calculated according to the equation: ∆𝐺 = −𝑅𝑇 ln 𝐾 = ∆𝐻 − 𝑇∆𝑆 Equation 1 Where R is the gas constant and T is the absolute temperature. In order to determine these thermodynamic parameters, the ligands are usually titrated into the oligonucleotide solution and the heat response is recorded. These heat values are usually corrected for the heat of ligand dilution. The representation of the heat values as a function of the molar ratio between the ligand and the Gquadruplex is known as the binding isotherm. Finally, fitting the binding isotherms to the proposed model allows the thermodynamic parameters discussed above to be obtained. This technique allows the interaction of a wide variety of ligands to be studied. There are examples of porphyrins (TMPyP4 and its derivatives) [26, 39, 48, 49, 148], quinoline alkaloids (berberine, palmatine or sanguinarine) [46], oxazole containing macrocycles (telomestatin and its derivatives) [58], distamycin A [109, 116, 149], actinomycin D [150], bisaryldiketene derivatives [95], triazatruxene derivatives [45, 49] and quindoline derivatives [55]. For G-quadruplex-forming sequences, most of the work has been carried out using sequences based on human telomeric DNA although sequences based on oncogenes such as cmyc, c-kit or bcl-2 have also been studied. The work of Arora and coworkers is an example of the possibilities of ITC [26]. There, the interaction of TMPyP4 with three different intramolecular G-quadruplex-forming sequences (c-myc, c-kit and telomeric DNA) was studied (see Figure 6). In all three cases the DNA:ligand stoichiometry was 1:3 and two different binding sites were identified. The strong binding site allowed the interaction of only one ligand molecule with an affinity constant of approximately 107 M-1 (end-stacking) which was significantly smaller in the case of telomeric DNA. The authors suggested that the differences in affinity constants could be 13 explained by the parallel structures of the c-myc and c-kit G-quadruplexes in contrast to the antiparallel structure of the telomeric DNA. The weak binding site allowed the interaction of two ligands with an affinity constant was 106 M-1 (external binding). Figure 6 near here A different application of ITC is presented in the work by Trotta et al. [116]. In that case, d(TGGGGT)4 was targeted with different ligands obtained from a virtual screening experiment. As the water solubility of the ligands was poor, ITC titration could not be carried out in the usual manner and competition experiments using distamycin A were carried out (in this case the ability of distamycin A to bind the G-quadruplex in the presence of another compound was measured). It was shown that if the ligand considered bound to the G-quadruplex more weakly than distamycin A did then the titration provided results similar to those obtained with a classic ITC experiment (no significant changes in the thermodynamic parameters). However, if the ligand bound to the G-quadruplex more strongly than distamycin A did then the results obtained in the titration were totally different and it was not possible to obtain a binding isotherm. So, these competition experiments allowed the relative strength of the interaction between the G-quadruplex structure and the ligands to be determined but there was a lack of thermodynamic information (stoichiometries, binding constants, etc) Competitive dialysis Competition dialysis is another useful quantitative tool for discovering structure-selective compounds that recognize particular nucleic acid structures [151, 152]. A competition dialysis experiment is based on dialysis of an array of nucleic acid structures against a common test ligand solution. Dialysis experiments involve a physical process where one or more solutes diffuse through a semi-permeable membrane. The flow of molecules across a membrane depends on the relation between their size and the membrane pore size: smaller molecules can diffuse across the membrane easily whilst larger molecules may be locked inside the membrane. The most common uses of dialysis are the removal of impurities or changes in solution conditions, i.e., during the purification of nucleic acids and proteins. Competition dialysis is a development of the dialysis method that allows the equilibrium specificity of a test ligand to be studied for different nucleic acid structures. The simplest equilibrium dialysis assay consists of two chambers each one containing a known concentration of a G-quadruplex and a ligand, respectively, separated by a semi-permeable membrane in which. This membrane allows the ligand to pass from one chamber to the other but the G-quadruplex is not allowed through and so it is retained in one chamber. Thus, some molecules of the ligand will interact with the G-quadruplex structure and other molecules will remain free in solution. At equilibrium, the concentration of free ligand is the same in both chambers but in the G-quadruplex chamber the concentration will be higher due to the quantity of bound ligand forming the complex which depends on the affinity between the sequence considered and the ligand. In a competition dialysis test, a solution of the ligand is put in touch with a number of individual closed chambers that contain different DNA sequences. So, depending on the affinity between each sequence and the ligand, the concentration of ligand in the chamber will vary. A deeper insight into the theoretical and practical aspects of competition dialysis applied to the study of G-quadruplex–ligand interactions can be found in the Ragazzon review [13]. Recent examples of the application of competition dialysis to the study of G-quadruplex-ligand interactions using UV-vis molecular absorption or fluorescence spectroscopy as the detection method can be found in the literature [32, 41, 43, 62, 110, 123, 153-155]. For instance, Xue and coworkers [62] showed that a perylene-neomycin conjugate had the strongest preference for human telomeric DNA compared to other DNA sequences (duplex, G-quadruplexes, etc). In another recent example, Ferreira [110] compared the binding preference of some acridine and quindoline oligomers with different G-quadruplex forming sequences such as c-myc, blc-2 or the human telomeric sequence (among other DNA secondary structures such as duplexes or triplexes). Acridine 4-aminoproline oligomers showed selective interaction with G-quadruplexes. In contrast, only the dimer quindoline ligand showed certain selectivity with the G-quadruplex of all the quindoline derivatives considered. Wang and coworkers [43] studied the interactions of different platinum-based complexes with the G-quadruplexes formed within the c-myc gene promoter. These ligands showed a preference to interact with the G-quadruplex structures compared to the duplex structures. The last example reported here corresponds to the work of Alcaro et al. [154] in which the G-quadruplex structure formed by the human telomeric based sequence d[AG3(T2AG3)3] showed a clear preference for fluorenone derivatives substituted with a morpholino moiety compared to duplex and triplex DNA structures. 14 Electrophoresis and other instrumental techniques Gel electrophoresis experiments enable the separation of DNA molecules based on size and charge using an electric field. The gel (usually agarose or polyacrylamide) is placed in an electrophoresis chamber that is connected to a power source. When an electric current is applied, the larger molecules move slowly through the gel while the smaller molecules move faster and, in the case of the study of drug:DNA interactions, the distance moved by the free G-quadruplex structure and the bound structure may be different. The molecules are usually detected using spectroscopic methods (molecular fluorescence or UV-visible absorption) and a previous step of staining is usually required [156]. Some examples of other separation techniques that could be used to study G-quadruplex–ligand interactions include that in the work of Waigh and coworkers [157-160] using capillary electrophoresis. Despite not applying this technique to the study of Gquadruplex-ligand interactions, they showed the potential of the technique to identify and quantify binding between duplex structures and ligands. As is it possible to separate the oligonucleotides, this technique is also suited to carrying out competition experiments in which a ligand is added to a mixture of different oligonucleotides. Another separation technique that could be considered is chromatography. This technique can directly analyze the interaction of a ligand and G-quadruplex structure, or the binding a ligand (such as the ligand N-methyl mesoporphyrin IX (NMM)) to a resin which is able to selectively interact with G-quadruplex-forming sequences [161]. Finally, it is also worth to mention the application of electrochemical methods to study the interactions between ligands and Gquadruplex structures [162-166]. For instance, Jin et coworkers [166] studied the changes in the voltammetric behavior of daunoribicin when interacts with different oligonucleotides (including a sequence corresponding to the human telomeric DNA). The results clearly show that the interaction between the daunorubicin and the G-quadruplex structure caused a decrease in the voltammetric signal corresponding to the non-complexed daunorubicin. Biochemical methods In addition to the instrumental methods described, interaction between ligands and G-quadruplex DNA structures are also studied by means of several biochemical methods. In this section, some of them are briefly described. First, some of these methods derive from the polymerase chain reaction and one of the assays most frequently used is the PCR stop assay [167]. The PCR stop assay is used to study the interference of ligands with polymerase activity, because it is an easy way to study whether a ligand stabilizes the G-quadruplex. If bands of paused polymerase activity are found at guanine rich segments of electrophoresis assays on PCR products, the efficacy of the stabilization by the ligand is demonstrated and the intensity of these paused bands is proportional to the inhibition of the polymerase activity. Several examples of the application of this assay to study G-quadruplex-ligand interactions can be found in the literature [27, 78, 87, 94, 97, 142, 168, 169]. Recently, Wang [170] has used this method to study whether the sequence (5’-G3(T2AG3)3-3’) is stabilized by phenantroline platinum complexes. The non-detection of PCR products allowed confirmation of the interaction between ligand and DNA and, also, its relative strength Another method with a PCR step is the telomeric repeat amplification protocol (TRAP) assay [171, 172]. In this case, the enzymatic activity of telomerase is measured, which is directly proportional to the amount of TTAGGG repeat present. In the PCR step, two different primers are used. The first primer acts as a substrate for telomerase-mediated addition of the TTAGGG repeats and the second primer is used as a reverse primer for PCR. A gel electrophoresis experiment using polyacrylamide is performed with the resultant products. For instance, Hang et al. [153] used this assay to study the effects of berberine and its derivatives on telomerase activity. However, Reed and coworkers [173] reported that the TRAP assay overestimated the activity of small molecules in inhibiting the enzyme because they interfered in the PCR step of the assay. For this reason, they proposed a modified TRAP assay known as TRAP-LIG in which the ligand is removed prior to the final PCR amplification of the products. Examples of the application of this recently proposed method can also be found in the literature [37, 61, 71, 73, 74, 91, 93, 94, 174-177]. Another common method used to identify G-quadruplex-ligand interaction is the Taq polymerase stop assay described by Han [178]. In this case, a duplex between the primer and the template is obtained and then the samples are incubated with the drugs 15 considered. Finally, there is the Taq polymerase extension and the gel electrophoresis analysis. Palumbo [68] used this assay to clearly identify the interactions of model ligands (TMPyP4 and telomestatin) with the G-quadruplex structures formed in the hTERT promoter. Other examples of the application of this method are the work of Rubis [179], Sun [79], Fu [27], Ren [61] and Rezler [180]. It is also possible to identify binding sites using footprinting assays [181]. Other experiments such as MTT assays [64, 71, 78, 87, 179], cell cultures [74, 79], DNase [182] or luciferase [57, 87, 97] assays have also been employed to identify interactions between G-quadruplex structures and ligands. Conclusions We have analyzed a large number of manuscripts that report interactions between G-quadruplex and ligands. It can be seen that every year the number of manuscripts dealing with interactions between G-quadruplex structures and molecules of biomedical interest increases and with this the number of instrumental techniques that are used to carry out different tests also grows. For this reason we have summarized in Table 2 the main properties of each of the techniques paying a special attention to the most important experimental requirements and the type of information the techniques that provide. As can be seen in Table 2, each technique provides a certain type of information (stoichiometries, type and strength of the binding, number of binding sites, kinetics of the binding reaction, etc) but none of them provides complete knowledge of the Gquadruplex-ligand system. So it is clear that if we want to have overall knowledge of a G-quadruplex-ligand system, a combination of complementary techniques must be used. Certainly, the number of experimental techniques available to study Gquadruplex-ligand systems depends on the capabilities of each laboratory, but a clear increase in the number of techniques used in each study has been observed in the works analyzed over recent years. A straightforward explanation of this fact is that the number of collaborations between different groups (each expert in a specific technique) has also increased and thus the maximum possible quantity of information is obtained from the system considered. It is clear that all this effort is directed at obtaining a complete characterization of the interactions between the G-quadruplex structures (formed both at the end of the telomeres and at the promoter regions of oncogenes) with different families of ligands. All the knowledge obtained from the study of these systems is intended to lead us to be able to design more effective new drugs that could help us to overcome diseases such as cancer. Acknowledgements We acknowledge funding from both the Spanish government (CTQ2009-11572 and CTQ2010-20541-C03-01) and the Catalan government (2009 SGR 45). 16 References [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] [12] [13] [14] [15] [16] [17] [18] [19] [20] [21] [22] [23] [24] [25] [26] [27] [28] [29] Neidle S, Balasubramanian S, Eds. Quadruplex Nucleic Acids. Cambridge, UK: Royal Society of Chemistry 2006. Bloomfield VA, Crothers DM, Tinoco Jr I, Eds. Nucleic Acids: Structures, Properties and Functions. Sausalito, US: University Science Books 2000. Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res, 2007; 35: 406-413. Wu Y, Brosh RM. G-quadruplex nucleic acids and human disease. FEBS J., 2010; 277: 3470-3488. Baumann P, Ed. G-Quadruplex DNA. New York, US: Humana Press 2010. Fox KR, Ed. Drug-DNA Interaction Protocols. New York, US: Humana Press 2010. Neidle S. Methods for studying quadruplex nucleic acids. Methods, 2007; 43: 245-245. Ho PS. Methods to study nucleic acid structure. Methods, 2009; 47: 141-141. Rachwal PA, Fox KR. Quadruplex melting. Methods, 2007; 43: 291-301. Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods, 2007; 43: 324-331. Webba da Silva M. NMR methods for studying quadruplex nucleic acids. Methods, 2007; 43: 264-277. Redman JE. Surface plasmon resonance for probing quadruplex folding and interactions with proteins and small molecules. Methods, 2007; 43: 302-312. Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods, 2007; 43: 313-323. Lee H-T, Olsen CM, Waters L, Sukup H, Marky LA. Thermodynamic contributions of the reactions of DNA intramolecular structures with their complementary strands. Biochimie, 2008; 90: 1052-1063. Rosu F, De Pauw E, Gabelica V. Electrospray mass spectrometry to study drug-nucleic acids interactions. Biochimie, 2008; 90: 1074-1087. Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, De Cian A, Mergny JL, Teulade-Fichou MP. Ligands playing musical chairs with G-quadruplex DNA: A rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie, 2008; 90: 1207-1223. Shirude PS, Balasubramanian S. Single molecule conformational analysis of DNA G-quadruplexes. Biochimie, 2008; 90: 1197-1206. Neidle S, Parkinson GN. Quadruplex DNA crystal structures and drug design. Biochimie, 2008; 90: 1184-1196. Murat P, Singh Y, Defrancq E. Methods for investigating G-quadruplex DNA/ligand interactions. Chem Soc Rev, in press (DOI: 10.1039/C1CS15117G). Sun H, Xiang J, Zhang Y, Xu G, Xu L, Tang Y. Spectroscopic studies of the interaction between methylene blue and G-quadruplex. Chinese Science Bulletin, 2006; 51: 1687-1692. Sun H, Tang Y, Xiang J, et al. Spectroscopic studies of the interaction between quercetin and G-quadruplex DNA. Bioorg Med Chem Lett, 2006; 16: 3586-3589. Wei C, Jia G, Yuan J, Feng Z, Li C. A spectroscopic study on the interactions of porphyrin with G-quadruplex DNAs. Biochemistry, 2006; 45: 6681-6691. Dincalp H, Avcibasi N, Icli S. Spectral properties and G-quadruplex DNA binding selectivities of a series of unsymmetrical perylene diimides. Journal of Photochemistry and Photobiology A: Chemistry, 2007; 185: 112. Barbieri CM, Srinivasan AR, Rzuczek SG, Rice JE, LaVoie EJ, Pilch DS. Defining the mode, energetics and specificity with which a macrocyclic hexaoxazole binds to human telomeric G-quadruplex DNA. Nucleic Acids Res, 2007; 35: 3272-3286. Galezowska E, Masternak A, Rubis B, et al. Spectroscopic study and G-quadruplex DNA binding affinity of two bioactive papaverine-derived ligands. International Journal of Biological Macromolecules, 2007; 41: 558-563. Arora A, Maiti S. Effect of loop orientation on quadruplex - TMPyP4 interaction. J Phys Chem B, 2008; 112: 8151-8159. Fu B, Zhang D, Weng X, et al. Cationic metal-corrole complexes: Design, synthesis, and properties of guaninequadruplex stabilizers. Chemistry, 2008; 14: 9431-9441. Mei WJ, Wei XY, Liu YJ, Wang B. Studies on the interactions of a novel ruthenium(II) complex with Gquadruplex DNA. Transition Metal Chemistry, 2008; 33: 907-910. Wei C, Han G, Jia G, Zhou J, Li C. Study on the interaction of porphyrin with G-quadruplex DNAs. Biophys Chem, 2008; 137: 19-23. 17 [30] [31] [32] [33] [34] [35] [36] [37] [38] [39] [40] [41] [42] [43] [44] [45] [46] [47] [48] [49] [50] [51] [52] [53] [54] del Toro M, Bucek P, Aviñó A, et al. Targeting the G-quadruplex-forming region near the P1 promoter in the human BCL-2 gene with the cationic porphyrin TMPyP4 and with the complementary C-rich strand. Biochimie, 2009; 91: 894-902. del Toro M, Gargallo R, Eritja R, Jaumot J. Study of the interaction between the G-quadruplex-forming thrombin-binding aptamer and the porphyrin 5,10,15,20-tetrakis-(N-methyl-4-pyridyl)-21,23H-porphyrin tetratosylate. Anal Biochem, 2008; 379: 8-15. Dik-Lung M, Che CM, Yan SC. Platinum(II) complexes with dipyridophenazine ligands as human telomerase inhibitors and luminescent probes for G-quadruplex DNA. J Am Chem Soc, 2009; 131: 1835-1846. Chen M, Song G, Wang C, Hu D, Ren J, Qu X. Small-molecule selectively recognizes human telomeric Gquadruplex DNA and regulates its conformational switch. Biophys J, 2009; 97: 2014-2023. Zhang H, Xiao X, Wang P, et al. Conformational conversion of DNA G-quadruplex induced by a cationic porphyrin. Spectrochim Acta A Mol Biomol Spectrosc, 2009; 74: 243-247. Arora A, Maiti S. Stability and molecular recognition of quadruplexes with different loop length in the absence and presence of molecular crowding agents. J Phys Chem B, 2009; 113: 8784-8792. Wei C, Jia G, Zhou J, Han G, Li C. Evidence for the binding mode of porphyrins to G-quadruplex DNA. Phys Chem Chem Phys, 2009; 11: 4025-4032. Bhattacharya S, Chaudhuri P, Jain AK, Paul A. Symmetrical bisbenzimidazoles with benzenediyl spacer: The role of the shape of the ligand on the stabilization and structural alterations in telomeric g-quadruplex dna and telomerase inhibition. Bioconjug Chem, 2010; 21: 1148-1159. Dutikova YV, Borisova OF, Shchyolkina AK, et al. 5,10,15,20-Tetra-(N-methyl-3-pyridyl)porphyrin destabilizes the antiparallel telomeric quadruplex d(TTAGGG)4. Mol Biol, 2010; 44: 823-831. Nagesh N, Buscaglia R, Dettler JM, Lewis EA. Studies on the site and mode of TMPyP4 interactions with Bcl-2 promoter sequence G-quadruplexes. Biophys J, 2010; 98: 2628-2633. Wei C, Wang J, Zhang M. Spectroscopic study on the binding of porphyrins to (G4T4G4)4 parallel Gquadruplex. Biophys Chem, 2010; 148: 51-55. Shi S, Geng X, Zhao J, et al. Interaction of [Ru(bpy)2(dppz)]2+ with human telomeric DNA: Preferential binding to G-quadruplexes over i-motif. Biochimie, 2010; 92: 370-377. Lee HM, Chan DSH, Yang F, et al. Identification of natural product Fonsecin B as a stabilizing ligand of c-myc G-quadruplex DNA by high-throughput virtual screening. Chem Commun, 2010; 46: 4680-4682. Wang P, Leung CH, Ma DL, Yan SC, Che CM. Structure-based design of platinum(II) Complexes as c-myc oncogene Down-regulators and luminescent probes for G-quadruplex DNA. Chemistry, 2010; 16: 6900-6911. Yaku H, Murashima T, Miyoshi D, Sugimoto N. Anionic phthalocyanines targeting G-quadruplexes and inhibiting telomerase activity in the presence of excessive DNA duplexes. Chem Commun, 2010; 46: 57405742. Petraccone L, Fotticchia I, Cummaro A, et al. The triazatruxene derivative azatrux binds to the parallel form of the human telomeric G-quadruplex under molecular crowding conditions: Biophysical and molecular modeling studies. Biochimie, 2011; 93: 1318-1327 Bhadra K, Kumar GS. Interaction of berberine, palmatine, coralyne, and sanguinarine to quadruplex DNA: A comparative spectroscopic and calorimetric study. BiochimBiophys Acta, 2011; 1810: 485-496. Jaumot J, Eritja R, Gargallo R. Chemical equilibria studies using multivariate analysis methods. Anal Bioanal Chem, 2011; 399: 1983-1997. Bhattacharjee AJ, Ahluwalia K, Taylor S, et al. Induction of G-quadruplex DNA structure by Zn(II) 5,10,15,20tetrakis(N-methyl-4-pyridyl)porphyrin. Biochimie, 2011; 93: 1297-1309. Cummaro A, Fotticchia I, Franceschin M, Giancola C, Petraccone L. Binding properties of human telomeric quadruplex multimers: A new route for drug design. Biochimie, 2011; 93: 1392-1400. Dash J, Waller ZAE, Pantos GD, Balasubramanian S. Synthesis and binding studies of novel diethynyl-pyridine amides with genomic promoter DNA G-quadruplexes. Chemistry, 2011; 17: 4571-4581. Cian AD, Mergny JL. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res, 2007; 35: 2483-2493. Sun H, Xiang J, Liu Y, Li L, Li Q, Xu G, Tang Y. A stabilizing and denaturing dual-effect for natural polyamines interacting with G-quadruplexes depending on concentration. Biochimie, 2011; 93: 1351-1356. Sun J, An Y, Zhang L, Chen HY, Han Y, Wang YJ, Mao ZW, Ji LN. Studies on synthesis, characterization, and Gquadruplex binding of Ru(II) complexes containing two dppz ligands. J Inorg Biochem, 2011; 105: 149-154. Cogoi S, Xodo LE. Enhanced G4-DNA binding of 5,10,15,20 (N-propyl-4-pyridyl) porphyrin (TPrPyP4): A comparative study with TMPyP4. Chem Commun, 2010; 46: 7364-7366. 18 [55] [56] [57] [58] [59] [60] [61] [62] [63] [64] [65] [66] [67] [68] [69] [70] [71] [72] [73] [74] [75] [76] [77] [78] [79] Xu W, Tan JH, Chen SB, Hou JQ, Li D, Huang ZS, Gu LQ. Studies on the binding of 5-N-methylated quindoline derivative to human telomeric G-quadruplex. Biochem Biophys Res Commun, 2011; 406: 454-458. Mergny JL, Lacroix L. Analysis of Thermal Melting Curves. Oligonucleotides, 2003; 13: 515-537. Agarwal T, Roy S, Chakraborty TK, Maiti S. Selective targeting of G-quadruplex using furan-based cyclic homooligopeptides: Effect on c-MYC expression. Biochemistry, 2010; 49: 8388-8397. Pilch DS, Barbieri CM, Rzuczek SG, LaVoie EJ, Rice JE. Targeting human telomeric G-quadruplex DNA with oxazole-containing macrocyclic compounds. Biochimie, 2008; 90: 1233-1249. Cheng X, Liu X, Bing T, Cao Z, Shangguan D. General peroxidase activity of G-quadruplex-hemin complexes and its application in ligand screening. Biochemistry, 2009; 48: 7817-7823. Li H, Liu Y, Lin S, Yuan G. Spectroscopy probing of the formation, recognition, and conversion of a Gquadruplex in the promoter region of the bcl-2 oncogene. Chemistry, 2009; 15: 2445-2452. Ren L, Zhang A, Huang J, et al. Quaternary ammonium zinc phthalocyanine: Inhibiting telomerase by stabilizing G quadruplexes and inducing G-quadruplex structure transition and formation. ChemBioChem, 2007; 8: 775-780. Xue L, Ranjan N, Arya DP. Synthesis and spectroscopic studies of the aminoglycoside (Neomycin)-perylene conjugate binding to human telomeric DNA. Biochemistry, 2011; 50: 2838-2849. Masiero S, Trotta R, Pieraccini S, et al. A non-empirical chromophoric interpretation of CD spectra of DNA Gquadruplex structures. Org Biomol Chem, 2010; 8: 2683-2692. Nadai M, Doria F, Di Antonio M, et al. Naphthalene diimide scaffolds with dual reversible and covalent interaction properties towards G-quadruplex. Biochimie, 2011; 93: 1328-1340. Micheli E, Martufi M, Cacchione S, De Santis P, Savino M. Self-organization of G-quadruplex structures in the hTERT core promoter stabilized by polyaminic side chain perylene derivatives. Biophys Chem, 2010; 153: 4353. Linder J, Garner TP, Williams HEL, Searle MS, Moody CJ. Telomestatin: Formal total synthesis and cationmediated interaction of its seco-derivatives with G-quadruplexes. J Am Chem Soc, 2010; 133: 1044-1051. Garner TP, Williams HEL, Gluszyk KI, et al. Selectivity of small molecule ligands for parallel and anti-parallel DNA G-quadruplex structures. Org Biomol Chem, 2009; 7: 4194-4200. Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc, 2009; 131: 10878-10891. White EW, Tanious F, Ismail MA, et al. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys Chem, 2007; 126: 140-153. Jantos K, Rodriguez R, Ladame S, Shirude PS, Balasubramanian S. Oxazole-based peptide macrocycles: A new class of G-quadruplex binding ligands. J Am Chem Soc, 2006; 128: 13662-13663. Wu WB, Chen SH, Hou JQ, et al. Disubstituted 2-phenyl-benzopyranopyrimidine derivatives as a new type of highly selective ligands for telomeric G-quadruplex DNA. Org Biomol Chem, 2011; 9: 2975-2986. Li Q, Xiang J, Li X, et al. Stabilizing parallel G-quadruplex DNA by a new class of ligands: Two non-planar alkaloids through interaction in lateral grooves. Biochimie, 2009; 91: 811-819. Wang L, Wen Y, Liu J, Zhou J, Li C, Wei C. Promoting the formation and stabilization of human telomeric Gquadruplex DNA, inhibition of telomerase and cytotoxicity by phenanthroline derivatives. Org Biomol Chem, 2011; 9: 2648-2653. Romera C, Bombarde O, Bonnet R, et al. Improvement of porphyrins for G-quadruplex DNA targeting. Biochimie, 2011; 93: 1310-1317. Keating LR, Szalai VA. Parallel-stranded guanine quadruplex interactions with a copper cationic porphyrin. Biochemistry, 2004; 43: 15891-15900. Shinohara KI, Sannohe Y, Kaieda S, et al. A chiral wedge molecule inhibits telomerase activity. J Am Chem Soc, 2010; 132: 3778-3782. Waller ZAE, Sewitz SA, Hsu STD, Balasubramanian S. A small molecule that disrupts G-quadruplex DNA structure and enhances gene expression. J Am Chem Soc, 2009; 131: 12628-12633. Ma Y, Ou TM, Tan JH, et al. Quinolino-benzo-[5, 6]-dihydroisoquindolium compounds derived from berberine: A new class of highly selective ligands for G-quadruplex DNA in c-myc oncogene. Eur J Med Chem, 2011; 46: 1906-1913. Sun D, Liu WJ, Guo K, et al. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol Cancer Ther, 2008; 7: 880-889. 19 [80] [81] [82] [83] [84] [85] [86] [87] [88] [89] [90] [91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103] [104] Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res, 2006; 34: 2536-2549. Owen EA, Keniry MA. Exploring the binding of calothrixin A to the G-quadruplex from the c-myc oncogene promotor. Aust J Chem, 2009; 62: 1544-1549. Alzeer J, Luedtke NW. PH-mediated fluorescence and G-quadruplex binding of amido phthalocyanines. Biochemistry, 2010; 49: 4339-4348. Dembska A, Juskowiak B. The fluorescence properties and lifetime study of G-quadruplexes single- and double-labeled with pyrene. J Fluoresc, 2010; 20: 1029-1035. Wei C, Wang L, Jia G, Zhou J, Han G, Li C. The binding mode of porphyrins with cation side arms to (TG4T)4 Gquadruplex: Spectroscopic evidence. Biophys Chem, 2009; 143: 79-84. Largy E, Hamon F, Teulade-Fichou MP. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal Bioanal Chem, 2011: 400: 3419-3427. Tran PLT, Largy E, Hamon F, Teulade-Fichou MP, Mergny JL. Fluorescence intercalator displacement assay for screening G4 ligands towards a variety of G-quadruplex structures. Biochimie, 2011; 93: 1288-1296. Chan DSH, Yang H, Kwan MHT, et al. Structure-based optimization of FDA-approved drug methylene blue as a c-myc G-quadruplex DNA stabilizer. Biochimie, 2011; 93: 1055-1064. De Cian A, Guittat L, Kaiser M, et al. Fluorescence-based melting assays for studying quadruplex ligands. Methods, 2007; 42: 183-195. Bertrand H, Bombard S, Monchaud D, Teulade-Fichou MP. A platinum-quinacridine hybrid as a G-quadruplex ligand. J Biol Inorg Chem, 2007; 12: 1003-1014. Sparapani S, Bellini S, Gunaratnam M, et al. Bis-guanylhydrazone diimidazo[1,2-a:1,2-c]pyrimidine as a novel and specific G-quadruplex binding motif. Chem Commun, 2010; 46: 5680-5682. Laronze-Cochard M, Kim YM, Brassart B, Riou JF, Laronze JY, Sapi J. Synthesis and biological evaluation of novel 4,5-bis(dialkylaminoalkyl)-substituted acridines as potent telomeric G-quadruplex ligands. Eur J Med Chem, 2009; 44: 3880-3888. Musetti C, Lucatello L, Bianco S, Krapcho AP, Cadamuro SA, Palumbo M, Sissi C. Metal ion-mediated assembly of effective phenanthroline-based G-quadruplex ligands. Dalton Trans, 2009: 3657-3660. Chen Z, Zheng KW, Hao YH, Tan Z. Reduced or diminished stabilization of the telomere G-quadruplex and inhibition of telomerase by small chemical ligands under molecular crowding condition. J Am Chem Soc, 2009; 131: 10430-10438. Ma Y, Ou TM, Tan JH, et al. Synthesis and evaluation of 9-O-substituted berberine derivatives containing azaaromatic terminal group as highly selective telomeric G-quadruplex stabilizing ligands. Bioorg Med Chem Lett, 2009; 19: 3414-3417. Peng D, Tan JH, Chen SB, Ou TM, Gu LQ, Huang ZS. Bisaryldiketene derivatives: A new class of selective ligands for c-myc G-quadruplex DNA. Bioorg Med Chem, 2010; 18: 8235-8242. Stefan L, Guedin A, Amrane S, et al. DOTASQ as a prototype of nature-inspired G-quadruplex ligand. Chem Commun, 2011; 47: 4992-4994. Wang XD, Ou TM, Lu YJ, et al. Turning off transcription of the bcl-2 gene by stabilizing the bcl-2 promoter quadruplex with quindoline derivatives. J Med Chem, 2010; 53: 4390-4398. Bejugam M, Gunaratnam M, Müller S, Sanders DA, Sewitz S, Fletcher JA, Neidle S, Balasubramanian S. Targeting the c-Kit promoter g-quadruplexes with 6-substituted indenoisoquinolines. ACS Med Chem Lett, 2010; 1: 306-310. Green JJ, Ladame S, Ying L, Klenerman D, Balasubramanian S. Investigating a quadruplex - Ligand interaction by unfolding kinetics. J Am Chem Soc, 2006; 128: 9809-9812. Jena PV, Shirude PS, Okumus B, et al. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. J Am Chem Soc, 2009; 131: 12522-12523. Pothukuchy A, Mazzitelli CL, Rodriguez ML, et al. Duplex and quadruplex DNA binding and photocleavage by trioxatriangulenium ion. Biochemistry, 2005; 44: 2163-2172. Zhang XF, Zhang HJ, Xiang JF, et al. The binding modes of carbazole derivatives with telomere G-quadruplex. J Mol Struct, 2010; 982: 133-138. Bieri M, Kwan AH, Mobli M, King GF, MacKay JP, Gooley PR. Macromolecular NMR spectroscopy for the nonspectroscopist: Beyond macromolecular solution structure determination. FEBS J, 2011; 278: 704-715. Kwan AH, Mobli M, Gooley PR, King GF, MacKay JP. Macromolecular NMR spectroscopy for the nonspectroscopist. FEBS J, 2011; 278: 687-703. 20 [105] [106] [107] [108] [109] [110] [111] [112] [113] [114] [115] [116] [117] [118] [119] [120] [121] [122] [123] [124] [125] [126] [127] [128] Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH. NMR-Based model of a telomeraseinhibiting compound bound to G-quadruplex DNA. Biochemistry, 1998; 37: 12367-74. Rangan A, Fedoroff OY, Hurley LH. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J Biol Chem, 2001; 276: 4640-6. Kern JT, Thomas PW, Kerwin SM. The relationship between ligand aggregation and G-quadruplex DNA selectivity in a series of 3,4,9,10-perylenetetracarboxylic acid diimides. Biochemistry, 2002; 41: 1137911389. Gavathiotis E, Heald RA, Stevens MFG, Searle MS. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J Mol Biol, 2003; 334: 25-36. Pagano B, Virno A, Mattia CA, Mayol L, Randazzo A, Giancola C. Targeting DNA quadruplexes with distamycin A and its derivatives: An ITC and NMR study. Biochimie, 2008; 90: 1224-1232. Ferreira R, Artali R, Farrera-Sinfreu J, et al. Acridine and quindoline oligomers linked through a 4aminoproline backbone prefer G-quadruplex structures. Biochim Biophys Acta, 2011; 1810: 769-776. Shang Q, Xiang JF, Zhang XF, Sun HX, Li L, Tang YL. Fishing potential antitumor agents from natural plant extracts pool by dialysis and G-quadruplex recognition. Talanta, 2011; 85: 820-823. Dash J, Shirude PS, Hsu STD, Balasubramanian S. Diarylethynyl amides that recognize the parallel conformation of genomic promoter DNA G-quadruplexes. J Am Chem Soc, 2008; 130: 15950-15956. Haider SM, Neidle S, Parkinson GN. A structural analysis of G-quadruplex/ligand interactions. Biochimie, 2011; 93: 1239-1251. Martino L, Virno A, Pagano B, et al. Structural and thermodynamic studies of the interaction of distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J Am Chem Soc, 2007; 129: 16048-16056. Hounsou C, Guittat L, Monchaud D, et al. G-quadruplex recognition by quinacridines: A SAR, NMR, and biological study. ChemMedChem, 2007; 2: 655-666. Trotta R, De Tito S, Lauri I, et al. A more detailed picture of the interactions between virtual screeningderived hits and the DNA G-quadruplex: NMR, molecular modelling and ITC studies. Biochimie, 2011; 93: 1280-1287. Beck JL, Colgrave ML, Ralph SF, Sheil MM. Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom Rev, 2001; 20: 61-87. Brodbelt JS. Evaluation of DNA/Ligand Interactions by Electrospray Ionization Mass Spectrometry. Annu Rev Anal Chem, 2010; 3: 67-87. Yuan G, Zhang Q, Zhou J, Li H. Mass spectrometry of G-quadruplex DNA: Formation, recognition, property, conversion, and conformation. Mass Spectrom Rev, in press (DOI: 10.1002/mas.20315). Nagesh N, Krishnaiah A, Dhople VM, Sundaram CS, Jagannadham MV. Noncovalent interaction of Gquadruplex DNA with acridine at low concentration monitored by MALDI-TOF mass spectrometry. Nucleosides Nucleotides Nucleic Acids, 2007; 26: 303-315. Gabelica V. Determination of equilibrium association constants of ligand-DNA complexes by electrospray mass spectrometry. Methods Mol Biol, 2010; 613: 89-101. Gabelica V, Baker ES, Teulade-Fichou MP, De Pauw E, Bowers MT. Stabilization and structure of telomeric and c-myc region intramolecular G-quadruplexes: The role of central cations and small planar ligands. J Am Chem Soc, 2007; 129: 895-904. Rosu F, De Pauw E, Guittat L, et al. Selective interaction of ethidium derivatives with quadruplexes: An equilibrium dialysis and electrospray ionization mass spectrometry analysis. Biochemistry, 2003; 42: 1036110371. Casagrande V, Alvino A, Bianco A, Ortaggi G, Franceschin M. Study of binding affinity and selectivity of perylene and coronene derivatives towards duplex and quadruplex dma by ESI-MS. J Mass Spectrom, 2009; 44: 530-540. Li H, Zhang Q, Yuan G. Investigation of the formation and recognition of a dimeric G-quadruplex in the promoter of Bcl-2 proto-oncogene by electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom, 2010; 24: 393-395. Li H, Yuan G. Electrospray ionization mass spectrometry probing of formation and recognition of the gquadruplex in the proximal promoter of the human vascular endothelial growth factor gene. Rapid Commun Mass Spectrom, 2010; 24: 2030-2034. Amato J, Oliviero G, Borbone N, et al. Ligand binding to tetra-end-linked (TGGGGT)4 G-quadruplexes: an electrospray mass spectroscopy study. Nucleic Acids Symp Ser (Oxf), 2008; 52: 165-166. Gornall KC, Samosorn S, Tanwirat B, et al. A mass spectrometric investigation of novel quadruplex DNAselective berberine derivatives. Chem Commun, 2010; 46: 6602-6604. 21 [129] [130] [131] [132] [133] [134] [135] [136] [137] [138] [139] [140] [141] [142] [143] [144] [145] [146] [147] [148] [149] [150] [151] [152] [153] [154] Gornall KC, Samosorn S, Talib J, Bremner JB, Beck JL. Selectivity of an indolyl berberine derivative for tetrameric G-quadruplex DNA. Rapid Commun Mass Spectrom, 2007; 21: 1759-1766. Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM. Evaluation of binding of perylene diimide and benzannulated perylene diimide ligands to DNA by electrospray ionization mass spectrometry. J Am Soc Mass Spectrom, 2006; 17: 593-604. Zhou J, Yuan G. Specific recognition of human telomeric G-Quadruplex DNA with small molecules and the conformational analysis by ESI mass spectrometry and circular dichroism spectropolarimetry. Chemistry, 2007; 13: 5018-5023. Tera M, Iida K, Ikebukuro K, Seimiya H, Shin-Ya K, Nagasawa K. Visualization of G-quadruplexes by using a BODIPY-labeled macrocyclic heptaoxazole. Org Biomol Chem, 2010; 8: 2749-2755. Moses JE, Ritson DJ, Zhang F, et al. A click chemistry approach to C3 symmetric, G-quadruplex stabilising ligands. Org Biomol Chem, 2010; 8: 2926-2930. David WM, Brodbelt J, Kerwin SM, Wang Thomas P. Investigation of quadruplex oligonucleotide - Drug interactions by electrospray ionization mass spectrometry. Anal Chem, 2002; 74: 2029-2033. Rosu F, Gabelica V, Shin-Ya K, De Pauw E. Telomestatin-induced stabilization of the human telomeric DNA quadruplex monitored by electrospray mass spectrometry. Chem Commun, 2003; 9: 2702-2703. Bianco S, Musetti C, Waldeck A, Sparapani S, Seitz JD, Krapcho AP, Palumbo M, Sissi C. Bis-phenanthroline derivatives as suitable scaffolds for effective G-quadruplex recognition. Dalton Trans, 2010; 39: 5833-5841. Homola J, Yee SS, Gauglitz G. Surface plasmon resonance sensors: review. Sensor Actuat B-Chem, 1999; 54: 3-15. Karlsson R. SPR for molecular interaction analysis: A review of emerging application areas. J Mol Recognit, 2004; 17: 151-161. Murat P, Bonnet R, Van Der Heyden A, et al. Template-Assembled Synthetic G-Quadruplex (TASQ): A Useful System for Investigating the Interactions of Ligands with Constrained Quadruplex Topologies. Chemistry, 2010; 16: 6106-6114. Waller ZAE, Shirude PS, Rodriguez R, Balasubramanian S. Triarylpyridines: A versatile small molecule scaffold for G-quadruplex recognition. Chem Commun, 2008: 1467-1469. Drewe WC, Nanjunda R, Gunaratnam M, et al. Rational design of substituted diarylureas: A scaffold for binding to G-quadruplex motifs. J Med Chem, 2008; 51: 7751-7767. Li G, Huang J, Zhang M, et al. Bis(benzimidazole)pyridine derivative as a new class of G-quadruplex inducing and stabilizing ligand. Chem Commun, 2008: 4564-4566. Dixon IM, Lopez F, Tejera AM, et al. A G-quadruplex ligand with 10000-fold selectivity over duplex DNA. J Am Chem Soc, 2007; 129: 1502-1503. Nielsen MC, Borch J, Ulven T. Design, synthesis and evaluation of 4,7-diamino-1,10-phenanthroline Gquadruplex ligands. Bioorg Med Chem, 2009; 17: 8241-8246. Halder K, Chowdhury S. Quadruplex-coupled kinetics distinguishes ligand binding between G4 DNA motifs. Biochemistry, 2007; 46: 14762-14770. Pillet F, Romera C, Trevisiol E, et al. Surface plasmon resonance imaging (SPRi) as an alternative technique for rapid and quantitative screening of small molecules, useful in drug discovery. Sensor Actuat B-Chem, 2011; 157: 304-309. Salim NN, Feig AL. Isothermal titration calorimetry of RNA. Methods, 2009; 47: 198-205. Erra E, Petraccone L, Esposito V, et al. Interaction of porphyrin with G-quadruplex structures. Nucleosides Nucleotides Nucleic Acids, 2005; 24: 753-756. Pagano B, Mattia CA, Virno A, Randazzo A, Mayol L, Giancola C. Thermodynamic analysis of quadruplex DNAdrug interaction. Nucleosides Nucleotides Nucleic Acids, 2007; 26: 761-765. Hudson JS, Brooks SC, Graves DE. Interactions of actinomycin D with human telomeric G-quadruplex DNA. Biochemistry, 2009; 48: 4440-4447. Ren J, Chaires JB. Rapid screening of structurally selective ligand binding to nucleic acids. Methods Enzymol. 2001; 340: 99-108. Ragazzon PA, Garbett NC, Chaires JB. Competition dialysis: A method for the study of structural selective nucleic acid binding. Methods, 2007; 42: 173-182. Zhang WJ, Ou TM, Lu YJ, et al. 9-Substituted berberine derivatives as G-quadruplex stabilizing ligands in telomeric DNA. Bioorg Med Chem, 2007; 15: 5493-5501. Alcaro S, Artese A, Iley JN, et al. Rational design, synthesis, biophysical and antiproliferative evaluation of fluorenone derivatives with DNA G-quadruplex binding properties. ChemMedChem, 2010; 5: 575-583. 22 [155] [156] [157] [158] [159] [160] [161] [162] [163] [164] [165] [166] [167] [168] [169] [170] [171] [172] [173] [174] [175] [176] [177] [178] [179] [180] Granotier C, Pennarun G, Riou L, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res, 2005; 33: 4182-4190. Martin R. Gel Electrophoresis: Nucleic Acids. Oxford, UK: Garland Science 1996. Araya F, Skellern GG, Waigh RD. Quantification of binding data using capillary electrophoresis. Methods Mol Biol, 2010; 613: 71-88. Araya F, Huchet G, McGroarty I, Skellern GG, Waigh RD. Capillary electrophoresis for studying drug-DNA interactions. Methods, 2007; 42: 141-149. Hamdan II, Skellern GG, Waigh RD. Use of capillary electrophoresis in the study of ligand-DNA interactions. Nucleic Acids Res, 1998; 26: 3053-3058. McGroarty I, Hamdan II, Skellern GG, Waigh RD. Determination of sequence binding preference of ligands with DNA using free solution capillary electrophoresis. J Pharm Pharmacol, 2000; 52: 144. Smith JS, Johnson FB. Isolation of G-quadruplex DNA using NMM-sepharose affinity chromatography. Methods Mol Biol, 2010; 608: 207-221. Jin Y, Li H, Liu P. Label-free electrochemical selection of G-quadruplex-binding ligands based on structure switching. Biosens Bioelectron, 2010; 25: 2669-2674. Liu XQ, Jin Y, Wang Y, Qiao Y. Electrochemical selection of G-quadruplex-binding ligands based on structureswitching of telomeric DNA. Anal Methods, 2011; 3: 1270-1273. Yang Q, Nie Y, Zhu X, Liu X, Li G. Study on the electrocatalytic activity of human telomere G-quadruplexhemin complex and its interaction with small molecular ligands. Electrochim Acta, 2009; 55: 276-280. Zhang KJ, Liu WY. Electrochemical and spectroscopic study on the interaction between G-quadruplex DNA and (R)-/(S)-2-(5-fluorouracil-1-acetyl) amido-1, 5-dimethyl glutarate. Int J Electrochem Sc, 2011; 6: 40064015. Jin Y, Qiao Y. A label-free method for identifying electroactive G-quadruplex-binding ligand. Electrochem Commun, 2010; 12: 966-969. Jennerwein MM, Eastman A. A polymerase chain reaction-based method to detect cisplatin adducts in specific genes. Nucleic Acids Res, 1991; 19: 6209-6214. Gomez D, Lamarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Research, 2004; 32: 371-379. Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou JF. Stabilization of the c-myc gene promoter quadruplex by specific ligands' inhibitors of telomerase. Biochem Biophys Res Commun, 2004; 323: 802-808. Wang JT, Zheng XH, Xia Q, Mao ZW, Ji LN, Wang K. 1,10-Phenanthroline platinum(II) complex: A simple molecule for efficient G-quadruplex stabilization. Dalton Trans, 2010; 39: 7214-7216. Evison BJ, Phillips DR, Cutts SM. In vitro transcription assay for resolution of drug-DNA interactions at defined DNA sequences. Methods Mol Biol, 2010; 613: 207-222. Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science, 1994; 266: 2011-2015. Reed J, Gunaratnam M, Beltran M, Reszka AP, Vilar R, Neidle S. TRAP-LIG, a modified telomere repeat amplification protocol assay to quantitate telomerase inhibition by small molecules. Anal Biochem, 2008; 380: 99-105. Gunaratnam M, Greciano O, Martins C, et al. Mechanism of acridine-based telomerase inhibition and telomere shortening. Biochem Pharmacol, 2007; 74: 679-689. De Cian A, DeLemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J Am Chem Soc, 2007; 129: 1856-1857. Moorhouse AD, Haider S, Gunaratnam M, Munnur D, Neidle S, Moses JE. Targeting telomerase and telomeres: A click chemistry approach towards highly selective G-quadruplex ligands. Mol BioSyst, 2008; 4: 629-642. Guittat L, De Cian A, Rosu F, et al. Ascididemin and meridine stabilise G-quadruplexes and inhibit telomerase in vitro. Biochim Biophys Acta, 2005; 1724: 375-384. Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res, 1999; 27: 537-542. Rubis B, Kaczmarek M, Szymanowska N, et al. The biological activity of G-quadruplex DNA binding papaverine-derived ligand in breast cancer cells. Invest New Drugs, 2009; 27: 289-296. Rezler EM, Seenisamy J, Bashyam S, et al. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J Am Chem Soc, 2005; 127: 9439-9447. 23 [181] [182] Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Footprinting: A method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods, 2007; 42: 128-140. Hampshire AJ, Fox KR. The effects of local DNA sequence on the interaction of ligands with their preferred binding sites. Biochimie, 2008; 90: 988-998. 24 Figures and Tables Figure 1. Example of the titration of a ligand with a G-quadruplex structure monitored by UV-visible molecular absorption. a) Experimental data obtained upon titration of TMPyP4 porphyrin with k-ras, inset: fit of the experimental data to the proposed model at 445 nm, b) distribution diagram and c) spectra of the recovered species using multivariate analysis. Reprinted with permission from [47]. Copyright 2011. Springer. Figure 2. Comparison of the melting profiles obtained for the 22-mer c-myc quadruplex in the presence of different quantities of two furan-based cyclic homooligopeptides. a) Ligand 1, b) Ligand 2 at a concentration of 0 M (), 5 M (), 10 M () and 15 M () . Reprinted with permission from [57]. Copyright 2010. American Chemical Society. Figure 3. Example of NMR titration of the parallel-stranded G-quadruplex form of [d(TAG3G4G5TTA)]4 with TOTA. The upfield shift of G3, G4, and G5 can be observed from a ratio of 0:1 (only G-quadruplex) to a 5:1 ratio (complex). Reprinted with permission from [102]. Copyright 2005. American Chemical Society. Figure 4. Example of ESI-MS spectra obtained from a mixture of 100 M guanine-rich and 100 M cytosine-rich sequences. a) in presence of 5 M of dehydrocorydaline and b) in presence of 80 M of dehydrocorydaline. Reprinted with permission from [60]. Copyright 2009. John Wiley and Sons. Figure 5. Example of SPR sensorgrams and analysis of the interaction between TMPyP4 and an intermolecular G-quadruplex. a) SPR sensorgrams upon increasing the ligand concentration from 10 nM to 250 nM, b) Adsorption isotherm for the interaction of TMPyP4 and the intermolecular quadruplex and c) Scatchard binding plot of the interaction between TMPyP4 and the intermolecular quadruplex. Reprinted with permission from [140]. Copyright 2010. John Wiley and Sons. Figure 6. Example of ITC profiles (left) and fitting curves using two independent sites (right) for the binding of TMPyP4 to a) cmyc, b) c-kit, c) telomeric quadruplexes and d) d) duplex DNA. Reprinted with permission from [26]. Copyright 2008. American Chemical Society. Table 1. Quantitation of G-quadruplex-ligand interactions. Table 2. Summary of the different experimental techniques used to study G-quadruplex-ligand interactions. 25 Figure 1. Example of the titration of a ligand with a G-quadruplex structure monitored by UV-visible molecular absorption. a) Experimental data obtained upon titration of TMPyP4 porphyrin with k-ras, inset: fit of the experimental data to the proposed model at 445 nm, b) distribution diagram and c) spectra of the recovered species using multivariate analysis. Reprinted with permission from [47]. Copyright 2011. Springer. 26 Figure 2. Comparison of the melting profiles obtained for the 22-mer c-myc quadruplex in the presence of different quantities of two furan-based cyclic homooligopeptides. a) Ligand 1, b) Ligand 2 at a concentration of 0 M (), 5 M (), 10 M () and 15 M () . Reprinted with permission from [57]. Copyright 2010. American Chemical Society. 27 Figure 3. Example of NMR titration of the parallel-stranded G-quadruplex form of [d(TAG3G4G5TTA)]4 with TOTA. The upfield shift of G3, G4, and G5 can be observed from a ratio of 0:1 (only G-quadruplex) to a 5:1 ratio (complex). Reprinted with permission from [102]. Copyright 2005. American Chemical Society. 28 Figure 4. Example of ESI-MS spectra obtained from a mixture of 100 M guanine-rich and 100 M cytosine-rich sequences. a) in presence of 5 M of dehydrocorydaline and b) in presence of 80 M of dehydrocorydaline. Reprinted with permission from [60]. Copyright 2009. John Wiley and Sons. 29 Figure 5. Example of SPR sensorgrams and analysis of the interaction between TMPyP4 and an intermolecular G-quadruplex. a) SPR sensorgrams upon increasing the ligand concentration from 10 nM to 250 nM, b) Adsorption isotherm for the interaction of TMPyP4 and the intermolecular quadruplex and c) Scatchard binding plot of the interaction between TMPyP4 and the intermolecular quadruplex. Reprinted with permission from [140]. Copyright 2010. John Wiley and Sons. 30 Figure 6. Example of ITC profiles (left) and fitting curves using two independent sites (right) for the binding of TMPyP4 to a) cmyc, b) c-kit, c) telomeric quadruplexes and d) d) duplex DNA. Reprinted with permission from [26]. Copyright 2008. American Chemical Society. 31 Table 1. Quantitation of G-quadruplex-ligand interactions. Target DNA sequence human telomeric human telomeric Techniques Stoichiometry Kb (M-1) Reference Scatchard plot Scatchard plot 1:1 2:1 105 105 [37] [55] human telomeric Scatchard plot 1:1 105 – 106 [46] human telomeric Scatchard plot 107 [53] Triazatruxene derivative human telomeric Scatchard plot 1:1 106 [45] Oxazine750 TMPyP4 Pt(II) complexes Diethynyl-pyridine amide TMPyP4, complementary Crich strand TMPyP4 human telomeric TBA c-myc c-kit Scatchard plot Multivariate analysis Scatchard plot Scatchard plot 4:1 1:1 1:1 1:1 106 105 107 107 [33] [31] [43] [50] bcl-2 Multivariate analysis 2:1 (TMPyP4) 106.5 (TMPyP4) 107 (C-rich) [30] k-ras Multivariate analysis 3:1 1018.9 [47] Ligand Bisbenzimidazole Quindoline derivatives Isoquinoline alkaloids Ru(II) complexes 32 Table 2. Summary of the different experimental techniques used to study G-quadruplex-ligand interactions. Technique Experiment type Titration Molecular absorption Melting Titration Circular Dichroism Melting Titration Melting Fluorescence NMR FRET G4-FID Qualitative Titration Titration SPR Titration ITC Titration Gel Electrophoresis Competitive Dialysis Quantitative (stoichiometries, binding constants) Qualitative Semi-quantitative (Tm, H, S, …) Quantitative (stoichiometries, binding constants) Qualitative Semi-quantitative (Tm, H, S, …) Quantitative (stoichiometries, binding constants) Qualitative Qualitative Quantitative Structure MS Information Capillary - Advantages Limitations Simplicity Small amount of sample (M) Poor selectivity Few information of the binding mode Simplicity Small amount of sample (M) Most ligands are non chiral Few information of the binding mode Simplicity Small amount of sample (M) Small amount of sample (M) Allows knowing the binding position Simplicity Small amount of sample (M) Qualitative (binding mode, stoichiometries, …) Qualitative Quantitative (stoichiometries, binding constants) Quantitative (stoichiometries, binding constants, binding mode, kinetic rates) Quantitative (stoichiometries, binding constants) Qualitative Quantitative (binding constants) Qualitative 33 Allows knowing the binding position Structure of the complex Soft-ionization sources required Different ligands simultaneously Ligand needs to be fluorescent (or use fluorescent bases in the DNA) Needs fluorophore labeled oligos Compatibility of the oligonucleotide with the fluorescent probe High amount of sample High amount of sample Limitations regarding the media (no K+ or Na+) Real-time Multiple samples simultaneously Biotin labeled oligonucletides Any type of ligand can be employed High amount of sample Water solubility of the ligands Multiple sequences simultaneously Poor reproducibility Low Sensitivity Simplicity Multiple sequences simultaneously Size of the structures Time of analysis