Tree-based Models for Classification Problems
advertisement

11 – Tree-based Models for Classification Problems
11.1 – Classification Trees
In classification trees the goals is to a tree-based model that will classify observations
into one of g predetermined classes. The end result of a tree model can be viewed as a
series of conditional probabilities (posterior probabilities) of class membership given a
set of covariate values. For each terminal node we essentially have a probability
distribution for class membership, where the probabilities are of the form:
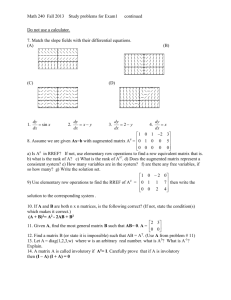
P (class i | x N A ) such that
~
g
P(class i | x N
i 1
~
A
) = 1.
Here, N A is a neighborhood defined by a set covariates/predictors, x .
~
The neighborhoods are found by a series of binary splits chosen to minimize the overall
“loss” of the resulting tree. For classification problems measuring overall “loss” can be a
bit complicated. One obvious method is to construct classification trees so that the
overall misclassification rate is minimized. In fact, this is precisely what the RPART
algorithm does by default. However in classification problems it is often times the case
we wish to incorporate prior knowledge about likely class membership. This knowledge
is represented by prior probabilities of an observation being from class i, which will
denote by i . Naturally the priors must be chosen in such a way that they sum to 1.
Other information we might want to incorporate into a modeling process is the cost or
loss incurred by classifying an object from class i as being from class j and vice versa.
With this information provided we would expect the resulting tree to avoid making the
most costly misclassifications on our training data set.
Some notation that is used by Therneau & Atkinson (1999) for determining the loss for a
given node A:
niA number of observations in node A from class i
ni number of observations in training set from class i
n total number of observations in the training set
n A number of observations in node A
n
i prior probability of being from class i (by default i i )
n
L(i, j ) = loss incurred by classifying a class i object as being from class j
( A) predicted class for node A
315
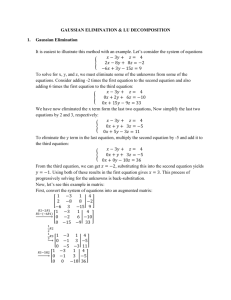
In general, the loss is specified as a matrix
L(1,2) L(1,3)
...
L(1, C )
0
L(2,1)
0
L(2,3)
...
L(2, C )
Loss Matrix L(3,1) L(3,2)
...
...
...
...
...
0
L(C 1, C )
...
L(C ,1) L(C ,2)
...
L(C , C 1)
0
By default this is a symmetric matrix with L(i, j ) L( j , i ) 1 for all i j .
Using the notation and concepts presented above the risk or loss at a node A is given by
C
n n
R( A) i L(i, ( A)) iA n A
i 1
ni n A
Example 11.1: Kyphosis Data
> library(rpart)
> data(kyphosis)
> attach(kyphosis)
> names(kyphosis)
[1] "Kyphosis" "Age"
"Number"
"Start"
> k.default <- rpart(Kyphosis~.,data=kyphosis)
> k.default
n= 81
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 81 17 absent (0.7901235 0.2098765)
2) Start>=8.5 62 6 absent (0.9032258 0.0967742)
4) Start>=14.5 29 0 absent (1.0000000 0.0000000) *
5) Start< 14.5 33 6 absent (0.8181818 0.1818182)
10) Age< 55 12 0 absent (1.0000000 0.0000000) *
11) Age>=55 21 6 absent (0.7142857 0.2857143)
22) Age>=111 14 2 absent (0.8571429 0.1428571) *
23) Age< 111 7 3 present (0.4285714 0.5714286) *
3) Start< 8.5 19 8 present (0.4210526 0.5789474) *
Sample Loss Calculations:
Root Node:
Terminal Node 3:
R(root) = .2098765*1*(17/17)*(81/81)*81 = 17
R(A) = .7901235*1*(8/64)*(81/19)*19 = 8
The actual tree is shown on the following page.
316
Suppose now we have prior beliefs that 65% of patients will not have Kyphosis (absent)
and 35% of patients will have Kyphosis (present).
> k.priors <-rpart(Kyphosis~.,data=kyphosis,parms=list(prior=c(.65,.35)))
> k.priors
n= 81
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 81 28.350000 absent (0.65000000 0.35000000)
2) Start>=12.5 46 3.335294 absent (0.91563089 0.08436911) *
3) Start< 12.5 35 16.453130 present (0.39676840 0.60323160)
6) Age< 34.5 10 1.667647 absent (0.81616742 0.18383258) *
7) Age>=34.5 25 9.049219 present (0.27932897 0.72067103) *
Sample Loss Calculations:
R(root) = .35*1*(17/17)*(81/81)*81 = 28.35
R(node 7) = .65*1*(11/64)*(81/25)*25 = 9.049219
The resulting tree is shown on the following page.
317
Finally suppose we wish to incorporate the following loss information.
Classification
L(i,j)
present absent
Actual present
0
4
absent
1
0
Note: In R the category orderings for the loss matrix are Z A in both dimensions.
This says that it is 4 times more serious to misclassify a child that actually has kyphosis
(present) as not having it (absent). Again we will use the priors from the previous model
(65% - absent, 35% - present).
> lmat <- matrix(c(0,4,1,0),nrow=2,ncol=2,byrow=T)
> k.priorloss <- rpart(Kyphosis~.,data=kyphosis,parms=list(prior=c(.65,.35),loss=lmat))
> k.priorloss
n= 81
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 81 52.650000 present (0.6500000 0.3500000)
2) Start>=14.5 29 0.000000 absent (1.0000000 0.0000000) *
3) Start< 14.5 52 28.792970 present (0.5038760 0.4961240)
6) Age< 39 15 6.670588 absent (0.8735178 0.1264822) *
7) Age>=39 37 17.275780 present (0.3930053 0.6069947) *
Sample Loss Calculations:
R(root) = .65*1*(64/64)*(81/81)*81 = 52.650000
R(node 7) = .65*1*(21/64)*(81/37)*37 = 17.275780
R(node 6) = .35*4*(1/17)*(81/15)*15 = 6.67058
The resulting tree is shown below on the following page.
318
Example 11.2 – Owl Diet
> owl.tree <- rpart(species~.,data=OwlDiet)
> owl.tree
n= 179
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 179 140 tiomanicus (0.14 0.21 0.13 0.089 0.056 0.22 0.16)
2) teeth_row>=56.09075 127 88 tiomanicus (0.2 0.29 0 0.13 0.079 0.31 0)
4) teeth_row>=72.85422 26
2 annandalfi (0.92 0.077 0 0 0 0 0) *
5) teeth_row< 72.85422 101 62 tiomanicus (0.0099 0.35 0 0.16 0.099 0.39 0)
10) teeth_row>=64.09744 49 18 argentiventer (0.02 0.63 0 0.27 0.02 0.061 0)
20) palatine_foramen>=61.23703 31 4 argentiventer (0.032 0.87 0 0.065 0 0.032 0)*
21) palatine_foramen< 61.23703 18
7 rajah (0 0.22 0 0.61 0.056 0.11 0) *
11) teeth_row< 64.09744 52 16 tiomanicus (0 0.077 0 0.058 0.17 0.69 0)
22) skull_length>=424.5134 7
1 surifer (0 0.14 0 0 0.86 0 0) *
23) skull_length< 424.5134 45
9 tiomanicus (0 0.067 0 0.067 0.067 0.8 0) *
3) teeth_row< 56.09075 52 24 whiteheadi (0 0 0.46 0 0 0 0.54)
6) palatine_foramen>=47.15662 25
1 exulans (0 0 0.96 0 0 0 0.04) *
7) palatine_foramen< 47.15662 27
0 whiteheadi (0 0 0 0 0 0 1) *
The function misclass.rpart constructs a confusion matrix for a classification tree.
319
> misclass.rpart(owl.tree)
Table of Misclassification
(row = predicted, col = actual)
1 2 3 4 5 6 7
annandalfi
24 2 0 0 0 0 0
argentiventer 1 27 0 2 0 1 0
exulans
0 0 24 0 0 0 1
rajah
0 4 0 11 1 2 0
surifer
0 1 0 0 6 0 0
tiomanicus
0 3 0 3 3 36 0
whiteheadi
0 0 0 0 0 0 27
Misclassification Rate =
0.134
Using equal prior probabilities for each of the groups.
> owl.tree2 <- rpart(species~.,data=OwlDiet,parms=list(prior=c(1,1,1,1,1,1,1)/7))
> misclass.rpart(owl.tree2)
Table of Misclassification
(row = predicted, col = actual)
1 2 3 4 5 6 7
annandalfi
24 2 0 0 0 0 0
argentiventer 1 27 0 2 0 1 0
exulans
0 0 24 0 0 0 1
rajah
0 4 0 11 1 2 0
surifer
0 1 0 1 9 5 0
tiomanicus
0 3 0 2 0 31 0
whiteheadi
0 0 0 0 0 0 27
Misclassification Rate = 0.145
> plot(owl.tree)
> text(owl.tree,cex=.6)
320
We can see that teeth row and palantine foramen figure prominently in the classification
rule. If you examine the group differences across these characteristics using comparative
boxplots this fact is not surprising.
Cross-validation is done in the usual fashion: we leave out a certain percentage of the
observations, develop a model from the remaining data, predict back the class of the
observations we set aside, calculate the misclassification rate, and then repeat this process
are number of times. The function crpart.cv leaves out p% of the data at a time to
perform the Monte Carlo cross-validation (MCCV) to estimate the APER.
> results = crpart.cv(owl.rpart,y=Owldiet$species,data=Owldiet,B=100)
> summary(results)
Min. 1st Qu. Median
Mean 3rd Qu.
Max.
0.1017 0.1695 0.2034 0.2193 0.2712 0.3559
> crpart.cv = function(fit,y,data,B=25,p=.333) {
n = length(y)
These options take extract any optional fitting
cv <- rep(0,B)
arguments used in the fitting process such as the
for (i in 1:B) {
prior probabilities, loss matrix, complexity
ss <- floor(n*p)
parameter (cp), and minsplit/minbucket info.
sam <- sample(1:n,ss)
temp <- data[-sam,]
fit2 <- rpart(formula(fit),data=temp,parms=fit$parms,control=fit$control)
ynew <- predict(fit2,newdata=data[sam,],type="class")
tab <- table(y[sam],ynew)
mc <- ss - sum(diag(tab))
cv[i] <- mc/ss
}
cv
}
321
11.2 - Bagging and Random Forests for Classification Trees
As with regression problems bagging (averaging trees built to bootstrap samples of the
training data) and random forests (building trees using random selected predictors at each
stage) can produce superior predictive performance.
Bagging (using the bagging() command in the ipred library)
> owl.bag = bagging(species~.,data=Owldiet,coob=T)
> owl.bag
Bagging classification trees with 25 bootstrap replications
Call: bagging.data.frame(formula=species ~ .,data = Owldiet, coob = T)
Out-of-bag estimate of misclassification error:
0.2123
> misclass(predict(owl.bag,newdata=Owldiet),Owldiet$species)
Table of Misclassification
(row = predicted, col = actual)
y
fit
annandalfi argentiventer exulans rajah surifer tiomanicus whiteheadi
annandalfi
25
0
0
0
0
0
0
argentiventer
0
37
0
0
0
0
0
exulans
0
0
24
0
0
0
0
rajah
0
0
0
16
0
0
0
surifer
0
0
0
0
10
0
0
tiomanicus
0
0
0
0
0
39
0
whiteheadi
0
0
0
0
0
0
28
Misclassification Rate =
0
The training cases are predicted perfectly with 0% misclassified, however the out-of-bag
estimate of the APER is .2123 or 21.23% misclassified.
Example 11.3: Italian Olive Oils
> mod = bagging(Area.name~.,data=O.area,coob=T)
> mod
Bagging classification trees with 25 bootstrap replications
Call: bagging.data.frame(formula = Area.name ~ ., data = O.area, coob = T)
Out-of-bag estimate of misclassification error:
0.0857
> misclass(predict(mod,newdata=O.area),O.area$Area.name)
Table of Misclassification
(row = predicted, col = actual)
y
fit
Calabria Coastal-Sardinia East-Liguria Inland-Sardinia North-Apulia Sicily South-Apulia Umbria West-Liguria
Calabria
56
0
0
0
0
0
0
0
0
Coastal-Sardinia
0
33
0
0
0
0
0
0
0
East-Liguria
0
0
50
0
0
0
0
0
0
Inland-Sardinia
0
0
0
65
0
0
0
0
0
North-Apulia
0
0
0
0
25
0
0
0
0
Sicily
0
0
0
0
0
36
0
0
0
South-Apulia
0
0
0
0
0
0
206
0
0
Umbria
0
0
0
0
0
0
0
51
0
West-Liguria
0
0
0
0
0
0
0
0
50
Misclassification Rate =
0
322
Again the training cases are predicted perfectly with 0% misclassified, however the outof-bag estimate of the APER is .0857 and 8.57% misclassified.
11.3 - Random Forests
Example 11.3 (cont’d): Italian Olive Oils
> area.rf = randomForest(Area.name~.,data=O.area,mtry=2,importance=T)
> area.rf
Call:
randomForest(formula = Area.name ~ ., data = O.area, mtry = 2,
importance = T)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 2
OOB estimate of
error rate: 4.72%
Confusion matrix:
Calabria Coastal-Sardinia East-Liguria Inland-Sardinia North-Apulia Sicily South-Apulia Umbria West-Liguria class.error
Calabria
54
0
0
0
0
0
2
0
0 0.03571429
Coastal-Sardinia
0
32
0
1
0
0
0
0
0 0.03030303
East-Liguria
0
0
46
0
0
0
0
1
3 0.08000000
Inland-Sardinia
0
0
0
65
0
0
0
0
0 0.00000000
North-Apulia
2
0
0
0
23
0
0
0
0 0.08000000
Sicily
4
0
0
0
3
23
6
0
0 0.36111111
South-Apulia
1
0
0
0
0
3
202
0
0 0.01941748
Umbria
0
0
0
0
0
0
0
51
0 0.00000000
West-Liguria
0
0
1
0
0
0
0
0
49 0.02000000
> area.rf = randomForest(Area.name~.,data=O.area,mtry=3,importance=T)
> area.rf
Call:
randomForest(formula = Area.name ~ ., data = O.area, mtry = 3,
importance = T)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 3
OOB estimate of error rate: 4.9%
> area.rf = randomForest(Area.name~.,data=O.area,mtry=4,importance=T)
> area.rf
Call:
randomForest(formula = Area.name ~ ., data = O.area, mtry = 4,
importance = T)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 4
OOB estimate of
error rate: 4.72%
323
Measures of Impurity (used to measure variable importance)
324
As you can see below the Gini Index decrease is used as a basis for one of the importance
measures for random forest models.
> area.rf$importance
palmitic
palmitoleic
strearic
oleic
linoleic
eicosanoic
linolenic
eicosenoic
Calabria Coastal-Sardinia East-Liguria Inland-Sardinia North-Apulia
Sicily South-Apulia
Umbria West-Liguria
0.13452872
0.073673892
0.02009836
0.063620230 0.219814164 0.02393492 0.035160949 0.18144189
0.07172358
0.12917763
0.297395878
0.24871128
0.258199312 0.294735044 0.12067000 0.349637005 0.26129882
0.05793994
0.05485467
0.008206269
0.02237659
0.020664404 0.002113947 0.17771535 0.048116657 0.09939278
0.01070815
0.11245305
0.282924624
0.23505554
0.355941504 0.230132746 0.06476381 0.165858075 0.69235145
0.24210970
0.32933668
0.508924377
0.45492602
0.495736641 0.316481124 0.14388783 0.224371017 0.48554629
0.34768423
0.23589078
0.031037995
0.02158836
0.007285309 0.012992940 0.02669415 0.008961566 0.05100809
0.16965593
0.07932334
0.016359483
0.03856755
0.012864521 0.041563009 0.10790249 0.014568810 0.01612817
0.38582919
0.28569336
0.189885202
0.47343418
0.166049624 0.307131929 0.25378467 0.080635695 0.39261139
0.23982349
palmitic
palmitoleic
strearic
oleic
linoleic
eicosanoic
linolenic
eicosenoic
MeanDecreaseAccuracy MeanDecreaseGini
0.07276472
28.36385
0.25472399
102.65161
0.04839498
23.73749
0.24516620
96.05318
0.33471962
95.28149
0.05194775
19.67056
0.06224676
42.75880
0.21391468
56.87026
> rfimp.class = function(rffit,measure=1,horiz=T) {
barplot(sort(rffit$importance[,measure]),horiz=horiz,xlab="Importance
Measure",main="Variable Importance")
}
> rfimp.class(area.rf,measure=11,horiz=F)
325
> rfimp.class(area.rf,measure=10,horiz=F)
The function below performs MCCV for a Random Forest model for classification.
> crf.cv =
function(fit,y,data,B=25,p=.333,mtry=fit$mtry,ntree=fit$ntree) {
n = length(y)
cv <- rep(0,B)
for (i in 1:B) {
ss <- floor(n*p)
sam <- sample(1:n,ss)
temp <- data[-sam,]
fit2 <- randomForest(formula(fit),data=temp,mtry=mtry,ntree=ntree)
ynew <- predict(fit2,newdata=data[sam,],type="class")
tab <- table(y[sam],ynew)
mc <- ss - sum(diag(tab))
cv[i] <- mc/ss
}
cv
}
> results = crf.cv(area.rf,y=O.area$Area.name,data=O.area)
> results
[1]
[8]
[15]
[22]
0.07894737
0.04736842
0.04210526
0.07368421
0.09473684
0.05789474
0.04736842
0.06315789
0.03684211
0.06842105
0.06842105
0.07894737
0.06842105 0.05263158 0.05263158 0.04210526
0.05263158 0.04210526 0.05789474 0.07894737
0.08947368 0.06315789 0.05789474 0.07368421
0.06315789
> summary(results)
Min. 1st Qu. Median
Mean 3rd Qu.
Max.
0.03684 0.05263 0.06316 0.06211 0.07368 0.09474
Proximities
These are another potentially useful byproduct of random forest fit. After a tree is grown,
run all of the data, both training and OOB, down the tree. If observations i and j are in the
same terminal node in a given tree increase their proximity by one. At the end, normalize
the proximities by dividing by the number of trees. The proximities are then returned in
an nxn matrix which can be visualized as a heat-map using the image()function.
326
> area.rf = randomForest(Area.name~.,data=O.area,mtry=4,importance=T,proximity=T)
> image(area.rf$proximity)
> title(main="Image Plot of Proximities for Olive Oils")
FYI ONLY: Multidimensional Scaling (MDS) is a method in which given set of
proximities (or distances = 1 – proximities) between a set of I observations in pdimensional space, a lower-dimensional representation (N = 2 or 3) is found so that the
distances between observations is preserved. These data can then be plotted in the lower
dimensional space so that interesting structure in the data can be observed graphically.
327
The following is taken from the Wikipedia page for MDS
The data to be analyzed is a collection of I objects on which a distance function is
defined,
δi,j := distance between i th and j th objects.
These distances are the entries of the dissimilarity matrix
The goal of MDS is, given Δ, to find vectors
for all
such that
,
where
is a vector norm. In classical MDS, this norm is the Euclidean distance but,
in a broader sense, it may be a metric or arbitrary distance function.
In other words, MDS attempts to find an embedding from the objects into RN such that
distances are preserved. If the dimension N is chosen to be 2 or 3, we may plot the
vectors xi to obtain a visualization of the similarities between the objects. Note that the
vectors xi are not unique. With the Euclidean distance for example, they may be
arbitrarily translated and rotated, since these transformations do not change the pairwise
distances
.
There are various approaches to determining the vectors xi. Usually, MDS is formulated
as an optimization problem, where
function, for example,
is found as a minimizer of some cost
A solution may then be found by numerical optimization techniques. For some
particularly chosen cost functions (e.g. squared error), minimizers can be stated
analytically in terms of matrix eigendecomposition, i.e. spectral decomposition of the
distance matrix. (End Wikipedia content)
The function MDSplot()will plot the results of a multidimensional scaling of the
distances formed by subtracting the proximities returned from a random forest fit from 1.
By default a two-dimensional plot is returned. The points are color-coded by the
categorical response variable.
328
> MDSplot(area.rf,O.area$Area.name,k=2)
> title(main="MDS Plot of Italian Olive Oils")
329
> MDSplot(area.rf,O.area$Area.name,k=3)
The MDS plots almost always look like this, which raises the question of their usefulness.
330
11.4 - Boosting for Classification Trees
Using function the boosting in the adabag library we can fit classification tree
models using the adaptive boosting method developed by Schapire & Freund. There are
other packages with boosting methods in them: mboost, ada, and gbm (which we used
for regression problems). For classification problems with non-binary/dichotomous
outcomes however, the boosting command in adabag appears to be only choice, and it
also seems to work fine when the response is dichotomous as well.
331
Example 11.3 (cont’d): Italian Olive Oils ~ classifying the growing area
> area.boost = boosting(Area.name~.,data=O.area,mfinal=200)
> summary(area.boost)
Length Class
Mode
formula
3
formula call
trees
200
-none- list
weights
200
-none- numeric
votes
5148
-none- numeric
prob
5148
-none- numeric
class
572
-none- character
importance
8
-none- numeric
> barplot(sort(area.boost$importance),main="Variable Importance")
> misclass(area.boost$class,O.area$Area.name)
Table of Misclassification
(row = predicted, col = actual)
y
fit
Calabria Coastal-Sardinia East-Liguria Inland-Sardinia North-Apulia Sicily South-Apulia Umbria
Calabria
56
0
0
0
0
0
0
0
Coastal-Sardinia
0
33
0
0
0
0
0
0
East-Liguria
0
0
50
0
0
0
0
0
Inland-Sardinia
0
0
0
65
0
0
0
0
North-Apulia
0
0
0
0
25
0
0
0
Sicily
0
0
0
0
0
36
0
0
South-Apulia
0
0
0
0
0
0
206
0
Umbria
0
0
0
0
0
0
0
51
West-Liguria
0
0
0
0
0
0
0
0
y
fit
West-Liguria
Calabria
0
Coastal-Sardinia
0
East-Liguria
0
Inland-Sardinia
0
North-Apulia
0
Sicily
0
South-Apulia
0
Umbria
0
West-Liguria
50
Misclassification Rate =
0
> plot(area.boost$trees[[1]])
> text(area.boost$trees[[1]])
> area.boost$weights[1]
[1] 1.035799
332
> plot(area.boost$trees[[180]])
> text(area.boost$trees[[180]],cex=.7)
> area.boost$weights[180]
[1] 1.067609
> area.boost$weights
[1]
[12]
[23]
[34]
[45]
[56]
[67]
[78]
[89]
[100]
[111]
[122]
[133]
[144]
[155]
[166]
[177]
[188]
[199]
1.0357992
0.9715957
0.9370240
0.8949037
0.8703193
1.1165439
0.9269494
0.8986684
0.8862763
1.4032175
0.8953054
1.0342709
0.7803303
1.2587555
1.0375580
1.0706908
1.0416944
0.7859464
1.1358410
1.1342587
0.9109128
0.9950483
1.1064092
1.1578435
0.9244610
0.9932527
0.8673571
0.8581576
1.0068071
1.0230568
0.8832552
1.1511980
0.8619396
1.0651710
1.0271797
1.1162135
1.0937164
1.0333415
1.0305158
1.0666677
0.8706232
0.8895321
0.8326164
1.0625838
1.0627201
1.1098483
0.7838882
1.0870706
1.0244511
1.3778578
0.9266824
0.8910515
0.9567762
1.0954239
0.9455997
0.8481177
1.0581825
0.9420402
0.8595090
1.0391414
1.0623264
0.9551809
1.0069796
0.9219371
0.9551105
1.0336974
1.0796687
1.0634951
1.0173223
1.0879095
0.9914885
0.8851813
1.0676094
0.8882285
0.9917211
1.0315845
0.9354693
0.8809439
1.0686684
1.0194853
1.0046468
1.0020261
1.0679582
0.8826356
0.9398476
1.0060252
0.9170549
0.8995054
0.9840809
0.9446243
1.0573716
1.1383453
1.0266454
0.8000069
1.3969888
1.0362868
0.8351552
0.8126341
1.1906942
1.0697031
1.1108903
0.9392506
1.0518340
1.0988167
0.9041655
1.1486299
1.1065162
1.0860789
1.1463685
1.1355113
0.8842932
0.9443035
1.1309866
0.9771788
1.1511997
1.0491668
0.8106182
0.9486628
1.1117184
1.2032414
1.0957845
1.2851301
1.0006903
0.9859894
1.1213506
1.0502042
1.2167139
1.0273235
1.0258271
0.9376967
0.9871757
1.0108042
0.9688051
1.0504185
0.9775952
0.9528626
0.9922303
0.9313945
1.0512727
0.9475067
1.0259476
0.8927899
0.9043262
0.8718052
0.9977561
1.1245070
0.9817287
1.3242475
0.8733040
1.0829109
0.9657496
1.0177336
1.0493857
1.4468478
0.9585049
0.9451793
1.0091305
1.0060947
0.9706092
1.0550020
1.3029086
0.8169246
1.0600536
0.8847374
0.9830194
1.0275357
0.9645878
0.8918500
0.9068563
1.0563784
1.0615412
1.0089787
1.0442129
0.9147896
1.2940220
1.0095361
0.9209586
1.1006079
1.0930421
0.9604656
0.9956170
0.9920956
1.0781610
0.9091424
0.9838000
0.8922770
1.1212727
1.0633402
0.8617204
1.0115171
1.1921323
0.9673191
0.9882869
1.2943851
1.3546597
1.0491384
0.9440773
1.0676094
0.9534185
0.9847003
> plot(area.boost$trees[[91]])
> text(area.boost$trees[[91]],cex=.7)
333
The original fit is potentially overfitting the training data so we should explore
this possibility by creating training/test sets from the original data or by
performing MCCV.
Test/Training Set Approach
>
>
>
>
>
sam = sample(1:572,100,replace=F)
Oarea.test = O.area[sam,]
Oarea.train = O.area[-sam,]
areaboost.train = boosting(Area.name~.,data=Oarea.train,mfinal=200)
misclass(areaboost.train$class,Oarea.train$Area.name)
Table of Misclassification
(row = predicted, col = actual)
y
fit
Calabria Coastal-Sardinia East-Liguria Inland-Sardinia North-Apulia Sicily South-Apulia Umbria
Calabria
42
0
0
0
0
0
0
0
Coastal-Sardinia
0
29
0
0
0
0
0
0
East-Liguria
0
0
44
0
0
0
0
0
Inland-Sardinia
0
0
0
55
0
0
0
0
North-Apulia
0
0
0
0
15
0
0
0
Sicily
0
0
0
0
0
32
0
0
South-Apulia
0
0
0
0
0
0
175
0
Umbria
0
0
0
0
0
0
0
41
West-Liguria
0
0
0
0
0
0
0
0
y
fit
West-Liguria
Calabria
0
Coastal-Sardinia
0
East-Liguria
0
Inland-Sardinia
0
North-Apulia
0
Sicily
0
South-Apulia
0
Umbria
0
West-Liguria
39
Misclassification Rate =
0
> predarea = predict(areaboost.train,newdata=Oarea.test)
> attributes(predarea)
$names
[1] "formula"
"votes"
"prob"
"class"
"confusion" "error"
> predarea$confusion
Observed Class
Predicted Class
Calabria Coastal-Sardinia East-Liguria Inland-Sardinia North-Apulia Sicily South-Apulia Umbria
Calabria
14
0
0
0
0
0
0
0
Coastal-Sardinia
0
4
0
0
0
0
0
0
East-Liguria
0
0
6
0
0
0
0
0
Inland-Sardinia
0
0
0
10
0
0
0
0
North-Apulia
0
0
0
0
9
0
0
0
Sicily
0
0
0
0
1
3
1
0
South-Apulia
0
0
0
0
0
1
30
0
Umbria
0
0
0
0
0
0
0
10
West-Liguria
0
0
0
0
0
0
0
0
Observed Class
Predicted Class
West-Liguria
Calabria
0
Coastal-Sardinia
0
East-Liguria
0
Inland-Sardinia
0
North-Apulia
0
Sicily
0
South-Apulia
0
Umbria
0
West-Liguria
11
> predarea$error
[1] 0.03 3% of the test cases were misclassified
334
> barplot(sort(area.boost$importance),horiz=F,
main="Variable Importances from AdaBoost")
> boost.cv = function(fit,y,data,p=.333,B=25,control=rpart.control())
{
n = length(y)
cv <- rep(0,B)
for (i in 1:B) {
ss <- floor(n*p)
sam <- sample(1:n,ss,replace=F)
temp <- data[-sam,]
fit2 <- boosting(formula(fit),data=temp,control=control)
ypred <- predict(fit2,newdata=data[sam,])
tab = ypred$confusion
mc <- ss - sum(diag(tab))
cv[i] <- mc/ss
}
cv
}
> results = boost.cv(area.boost,O.area$Area.name,data=O.area,B=100)
> summary(results)
Min. 1st Qu. Median
Mean 3rd Qu.
Max.
0.03158 0.05132 0.05789 0.06232 0.07368 0.10530
335
> Statplot(results)
There are three different weighting formulae available for use with the boosting function
in the adabag library. The argument coeflearn has the following options:
1
Breiman (default):
𝛼 = 2 ln (
Freund:
𝛼 = 𝑙𝑛 (
Zhu:
𝛼 = 𝑙𝑛 (
𝑒𝑡 −1
)
𝑒𝑡
𝑒𝑡 −1
)
𝑒𝑡
𝑒𝑡 −1
𝑒𝑡
) + ln(# 𝑜𝑓 𝑐𝑙𝑎𝑠𝑠𝑒𝑠 − 1)
How much the performance varies across these different weighting schemes can
be explored on an application by application basis, choosing the one with the
optimal performance as judged by MCCV or a test/training set approach.
For problems when the response is dichotomous other libraries with boosting
algorithms can be used. The function ada in the library of the same name will
perform discrete, real, or gentle boosting. Real and Gentle boosting do not use
misclassification error, but rather weight according to the estimated probabilities
y = 1 given the x’s. For example, the weights for Real Boosting are determined by
𝛼=
1
1 − 𝑝𝑚 (𝑥)
𝑙𝑛 (
)
2
𝑝𝑚 (𝑥)
336
where 𝑝𝑚 (𝑥) = 𝑃̂(𝑦 = 1|𝒙). For two class prediction problems it is probably
worth exploring a variety of these methods for a given problem to arrive an
optimal choice. My guess is the “optimal” choice, determined by some form of
validation, will vary from problem to problem.
Example 11.4: Cleveland Heart Disease Study
> diag.ada = ada(diag~.,data=Cleve.diag)
> diag.ada
Call:
ada(diag ~ ., data = Cleve.diag)
Loss: exponential Method: discrete
Iteration: 50
Final Confusion Matrix for Data:
Final Prediction
True value buff sick
buff 151
9
sick
16 120
Train Error: 0.084
Out-Of-Bag Error:
0.108
iteration= 41
Additional Estimates of number of iterations:
train.err1 train.kap1
44
44
> varplot(diag.ada)
337
> diag.adaboost = boosting(diag~.,data=Cleve.diag,mfinal=50)
> misclass(diag.adaboost$class,Cleve.diag$diag)
Table of Misclassification
(row = predicted, col = actual)
y
fit
buff sick
buff 160
0
sick
0 136
Misclassification Rate =
0
MCCV Results
> boost.results = boost.cv(diag.adaboost,Cleve.diag$diag,Cleve.diag)
> summary(boost.results)
Min. 1st Qu. Median
Mean 3rd Qu.
Max.
0.1327 0.1735 0.1939 0.1967 0.2245 0.2653
> ada.results = ada.cv(diag.ada,Cleve.diag$diag,Cleve.diag)
> summary(ada.results)
Min. 1st Qu. Median
Mean 3rd Qu.
Max.
0.1122 0.1429 0.1633 0.1694 0.1939 0.2347
The boosting methods in the ada outperform those in the adabag package for
this two class prediction problem.
The ada function also allows for easy prediction for a test set with a visualization
of the results.
> Cleve.test = Cleve.diag[250:296,]
> Cleve.train = Cleve.diag[1:249,]
> diag.train = ada(diag~.,data=Cleve.train)
> diag.test = addtest(diag.train,Cleve.test[,-14],Cleve.test[,14])
> diag.test
Call:
ada(diag ~ ., data = Cleve.train)
Loss: exponential Method: discrete
Iteration: 50
Final Confusion Matrix for Data:
Final Prediction
True value buff sick
buff 127
6
sick
14 102
Train Error: 0.08
Out-Of-Bag Error:
0.1
iteration= 48
Additional Estimates of number of iterations:
train.err1 train.kap1 test.errs2 test.kaps2
50
50
32
32
338
> plot(diag.test,test=T)
339
340
Misclassification Table for Tree-based Classification Models
misclass = function (tree)
{
temp <- table(predict(tree, type = "class"), tree$y)
cat("Table of Misclassification\n")
cat("(row = predicted, col = actual)\n")
print(temp)
cat("\n\n")
numcor <- sum(diag(temp))
numinc <- length(tree$y) - numcor
mcr <- numinc/length(tree$y)
cat(paste("Misclassification Rate = ", format(mcr, digits =
3)))
cat("\n")
}
This function should work for rpart, tree, bagging, and randomForests.
341
11.5 – Tree-based Models for Classification in JMP
To begin tree-based modeling for regression or classification problems in JMP select
Partition from the Modeling pull-out menu as shown below.
If the response (Y) is continuous it will fit a regression tree-based model and if the
response is nominal it will fit a classification tree-based model.
Decision tree - basically fits a
model like rpart in R.
Bootstrap Forest - can be used
to fit a bagged fit or a random
forest to improve the predictive
performance of the tree
models.
Boosted Tree - uses boosting
to improve the predictive
performance of tree for
dichotomous responses only!
Set a fraction of your training data to use as validation set, e.g.
.3333 to set aside 33.33% of your training data for validation
purposes.
342
Bagging and Random Forest Dialog Box in JMP
M = # of tree models to average.
Number of terms sampled per split = #
of variables randomly sampled at each
potential split (mtry in R) .
Bootstrap sample rate = proportion of
observations sampled for each
bootstrap.
Minimum Splits Per Tree = is the
minimum number of splits for each
tree
Maximum Splits Per Tree = maximum
tree size for any tree fit to a bootstrap
sample.
Minimum Size Split = smallest
number of observations per node
where further splitting would be
considered.
Early Stopping = will average less than
M trees if averaging more trees does
not improve validation statistic.
Multiple Fits over number of terms =
will fit random forests using the value
specified in Number of terms per
sampled split to Max Number of terms
and return the final fit will be the best
random forest obtained using values in
this range.
343